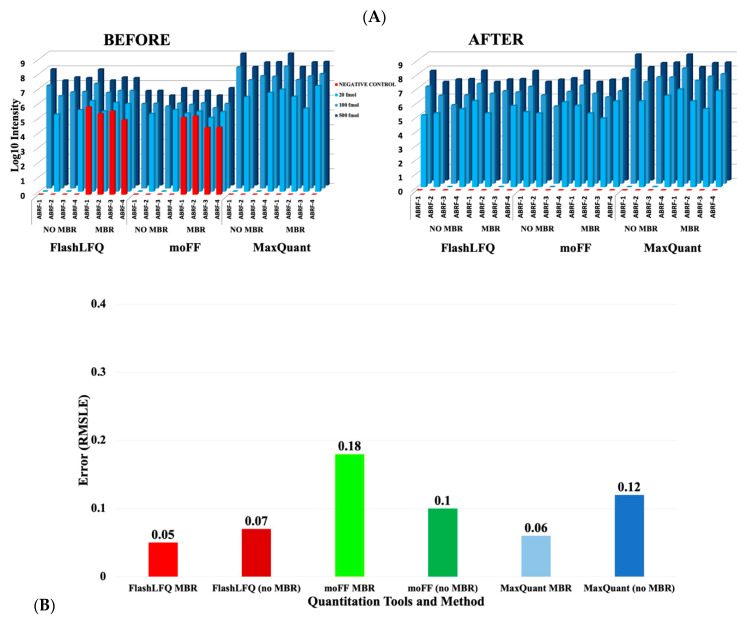
Figure 3.
(A) Effect of MBR after software version updates: The log10 values of the intensities (blue bars) from each of the four ABRF spiked-in proteins (ABRF-1: beta Galactosidase from E. coli, ABRF-2: Lysozyme from Gallus gallus, ABRF-3: amylase from Aspergillus, ABRF-4: protein G Streptococcus) were plotted. The results from prior versions of moFF (v1.2.1) and FlashLFQ (v0.1.99) (before) shows that MBR detects ABRF proteins (shown in red) in the negative control sample in both software. The results from the current versions of moFF (v2.0.2) and FlashLFQ (v1.0.3.0) implemented in Galaxy (after), shows that the MBR feature does not detect ABRF proteins in the negative control. (B) Accuracy of fold-change estimation: for evaluating the accuracy of quantified results, we estimated the fold change of the spiked-in proteins in the 500 fmol sample as compared to 100 fmol sample. The root mean squared log error (RMSLE) was calculated for fold change estimation. For this dataset, moFF with MBR displayed significantly higher RMSLE value, whereas FlashLFQ’s MBR performed similarly to MaxQuant’s MBR.