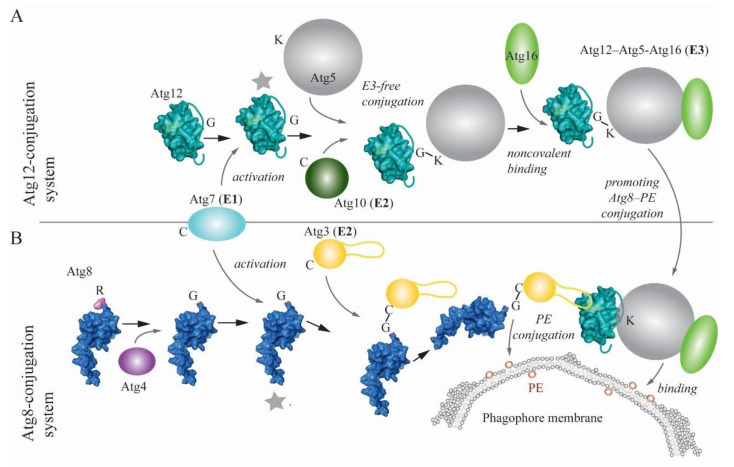
Figure 1.
Schematic representation of the Atg8- and Atg12-conjugation systems in yeast. (A) Atg12 is activated by Atg7, the E1-like (activating) enzyme, and then transferred to Atg10, the E2-like (conjugating) enzyme. The Atg12–Atg10 intermediate interacts with Atg5, where the conserved lysine residue is covalently conjugated to the Atg12 C terminus in the E3-free (ligase) reaction. The Atg12–Atg5 binds noncovalently to Atg16. The resulting Atg12–Atg5-Atg16 complex acts as the E3-like enzyme in the Atg8–PE conjugation reaction. (B) Atg8 enters the conjugation reaction unprimed, due to the presence of a C-terminal arginine. Atg4 primes Atg8 by removing this last residue, leaving a C-terminal glycine exposed. As in the case of Atg12, Atg7 activates the Atg8 ubiquitin-like (UBL) domain by C-terminal adenylation, and then transfers Atg8 to the catalytic cysteine in the active site of Atg3. The Atg12–Atg5-Atg16 complex (E3-like) interacts with the Atg8~Atg3 intermediate, where a long flexible loop of Atg3 binds to a hydrophobic cavity on the surface of Atg12. This interaction enhances ligation of the C-terminal Gly in Atg8 to PE on the phagophore membrane. Atg8 and Atg12 are visualized using the crystal structures PDB ID: 2KQ7 and PDB ID: 3W1S, respectively. Gray stars denote activated states of molecules.