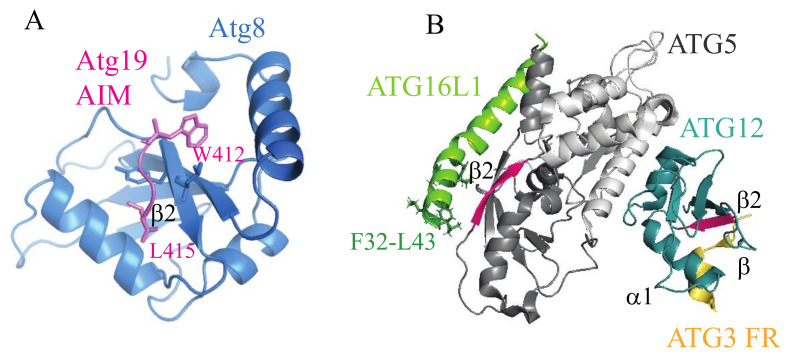
Figure 2.
Binding of the autophagy related 8 (Atg8)-interacting motif (AIM)/microtubule associated protein 1 light chain 3 (LC3)-interacting region (LIR) sequences to the ubiquitin-like folds in the autophagy machinery. (A) The Atg8 ubiquitin-like (UBL) folds in a complex with the Atg19 AIM peptide (PDB ID: 2ZPN). Hydrophobic amino acid residues in the AIM motif (WEEL) are inserted into the two hydrophobic cavities (W and L site) on the flanking surface areas of the β2 strand. The Atg19 AIM tetrapeptide and the Atg8 β2 strand form an intermolecular β-sheet, a secondary structure found in all canonical AIM/LIR-Atg8/LC3/GABA type A receptor-associated protein (GABARAP) interactions. (B) The ATG3-ATG12–ATG5-ATG16N complex (PDB ID: 4NAW) in ribbon representation. When ATG3 binds to ATG12, the β2 strand (pink) of ATG12 forms an intermolecular β sheet with the β strand (orange) of the AADM157 tetrapeptide in the disordered region of ATG3 (ATG3 FR). The UBL-A domain (dark gray) on ATG5 possesses the β2 strand (pink). It remains to be elucidated if the ATG5 β2 forms an intermolecular β sheet with the β strand of the LIR motifs in autophagy receptors. Note that the ATG16L1 helical region spanning amino acid residues F32-L43 forms an amphipathic helix that inserts into a lipid bilayer [28,29], instead of binding to ATG5 that is seen in the crystal structure in the absence of membranes. FR, flexible region.