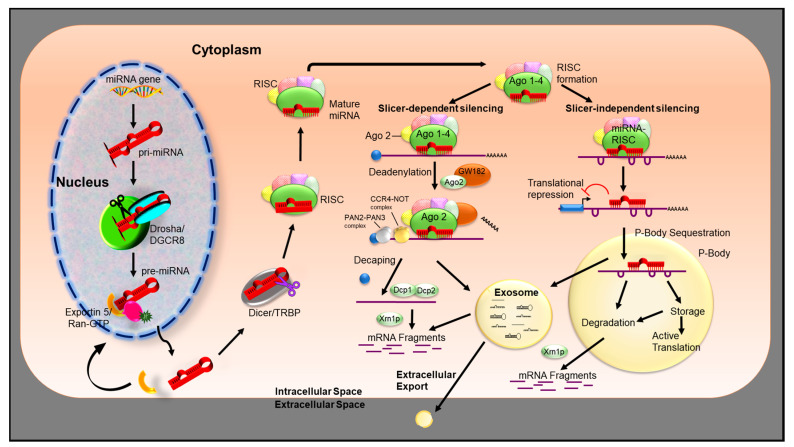
Figure 1.
miRNA biogenesis and mechanism of action. miRNAs are transcribed by RNA polymerase II or III, producing primary transcripts (pri-miRNA). The pri-miRNA is then cleaved by the Drosha and DGCR8 complex, producing precursor miRNA (pre-miRNA). Pre-miRNA assembles into a complex with Exportin-5 (XPO5) and Ran/GTP. Once in the cytoplasm, pre-miRNA is further processed by Dicer complex into mature miRNA. The passenger strand of mature miRNA is degraded, while the other strand is loaded into argonaute protein (Ago1–4) and incorporated into the RNA-induced silencing complex (RISC). Binding of RISC to its target mRNA results in degradation and/or translational repression of the target gene, by either slicer-independent or slicer-dependent silencing. When the miRNA is extensively base-paired, slicer-dependent silencing mechanisms proceed. GW182 family of proteins is recruited by AGO. GW182 interacts with PABPC, promoting efficient mRNA deadenylation through the recruitment of PAN2–PAN3 and CCR4–NOT complexes. Cleavage begins with the deadenylation of the mRNA to remove the poly (A) tail. Deadenylation promotes subsequent mRNA decapping and degradation by Xrn1p. Alternatively, degradation can occur via the exosome (vesicles with a size of 30 to 100nm, with 3′→5′ exonuclease activity). In slicer-independent silencing, multiple complementary sites with imperfect base-pairing create bulges in the RNA duplex propelling slicer-independent gene silencing mechanisms. miRNAs can repress translation determined by the target mRNA promoter. Alternatively, miRNA can repress translation indirectly by segregating mRNA into P-Bodies. Ultimately, mRNA can be isolated for storage or be targeted for decay via Xrn1p or exosome degradation.