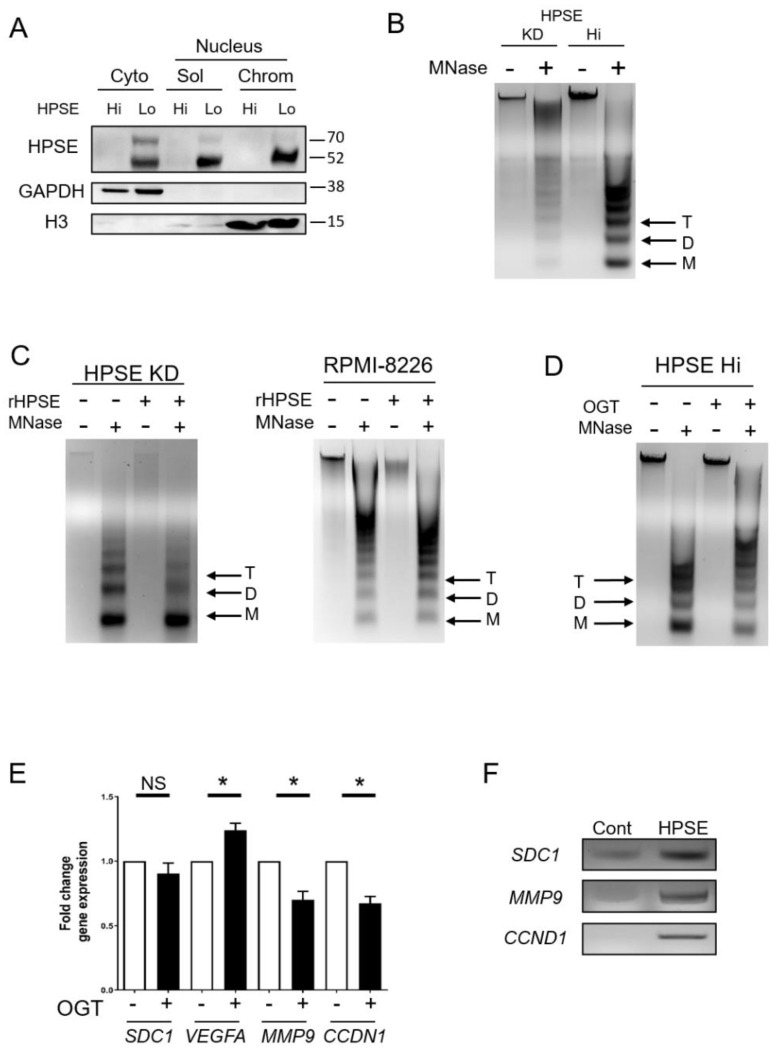
Figure 1.
Heparanase enhances chromatin accessibility. (A) To determine localization of heparanase, sequential cell fractionation of myeloma cells was used to isolate cytoplasmic (cyto), nucleus-soluble (sol), and nucleus-chromatin (chrom) fractions from CAG wild-type (HPSE Lo) and transfected CAG HPSE Hi cells prior to Western blot analysis. GAPDH and histone H3 (H3) were probed to assess the quality of the fractions. (B) Chromatin accessibility in CAG HPSE knockdown (KD) and HPSE Hi cells were assessed by sensitivity to micrococcal nuclease (MNase) digestion. Nuclei from KD and HPSE Hi cells were prepared and digested with 0.1 U/μL MNase and subjected to electrophoresis. Black arrows denote Tri (T), Di (D) and Mono (M)—nucleosomes. (C) Nuclei from CAG HPSE KD and RPMI-8226 myeloma cells were treated for 2 h with or without rHPSE before being subjected to Mnase digestion for chromatin accessibility. (D) CAG HPSE Hi cells were incubated with 20 µM of the HPSE inhibitor OGT2115 and chromatin accessibility was assessed following exposure of nuclei to MNase. (E) mRNA expression analysis by RT-PCR of syndecan-1 (SDC1), vascular endothelial growth factor A (VEGFA), matrix metalloproteinase-9 (MMP9) and cyclin D1 (CCDN1) in CAG HPSE Hi cells in the presence or absence of OGT2115, compared to respective control. * p < 0.05 by one-tailed, paired t-test. (F) Chromatin immunoprecipitation assay was performed on CAG HPSE Hi cells to analyze HPSE binding to the promoter regions of SDC1, MMP9 and CCND1. PCR was performed using primers probing the promoter regions of indicated genes after pulldown with an antibody control (Cont) or anti-heparanase (HPSE) and result was assessed by gel electrophoresis.