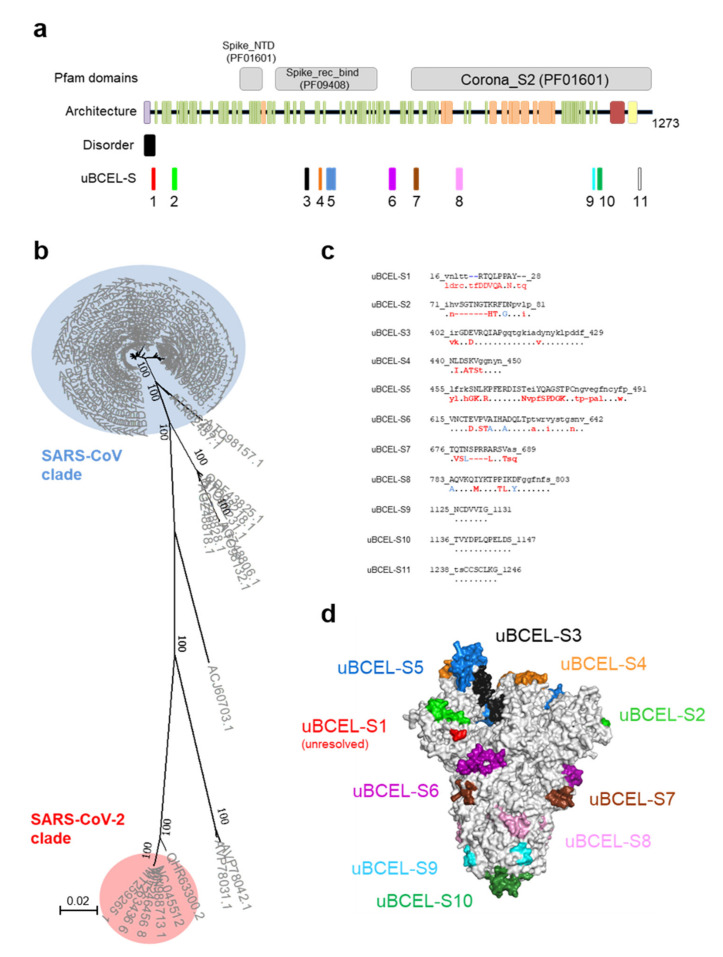
Figure 4.
Architecture and analysis of predicted uBCELs on spike S protein. (a) a graphical depiction of S protein with Pfam domains, architecture, disorder and uBCELs localization. Color blocks: Pfam domains (grey), alpha-helices (orange), beta-strand (green), transmembrane helices (yellow), signal peptide (purple), coiled-coils (red) and disordered regions (black). In addition, each uBCEL is shown in its own color; (b) phylogenetic tree of 89 non-redundant coronavirus S sequences calculated by the Neighbor-Joining method. Bootstrap values of 100 are indicated; (c) SARS-CoV-2 uBCEL-S sequences (including deletions) and changes observed in relation to SARS-CoV S protein: capital letters indicate epitopes, residues conserved in ≥90% sequences (dots), changed to unique option (≥90%, red), ambiguous changes (two or more residue option in >10% sequences, blue), and deletions (dashes); (d) structural mapping of uBCELs on the surface view of the modeled S protein homotrimer. Each uBCEL is depicted in the same colors as in Figure 4a. uBCEL-S11 falls out of the resolved section of the protein and therefore is not shown.