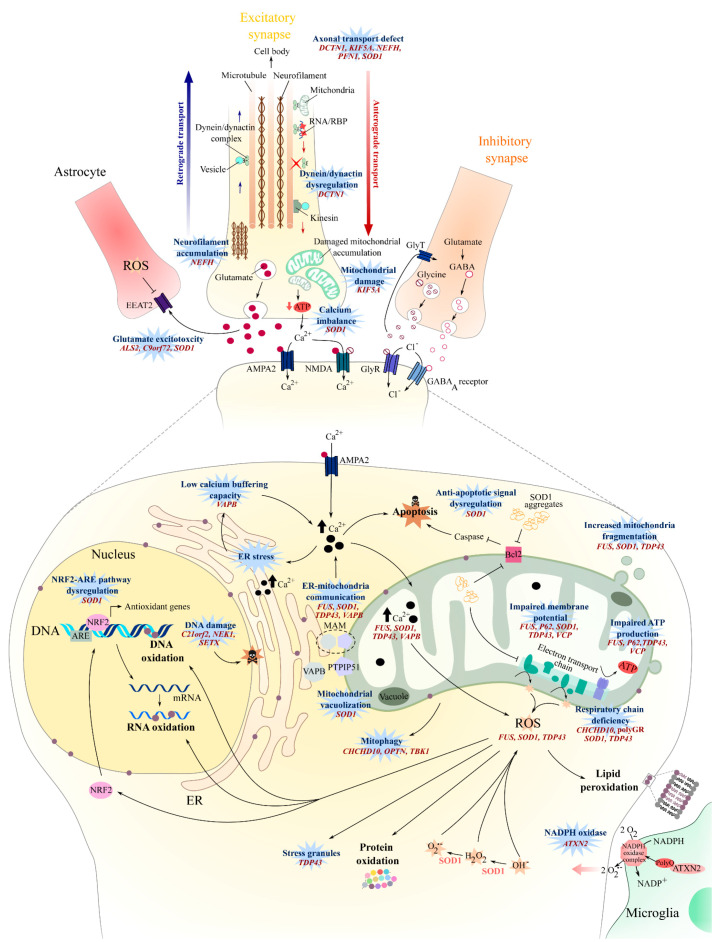
Figure 1.
Oxidative stress, mitochondrial dysfunction, axonal transport, and glutamate excitotoxicity in amyotrophic lateral sclerosis (ALS). An increase in oxidative stress can result from defects in detoxifying pathways. Such defects include the loss of SOD1 function, aberrant DNA damage repair machinery, or a decrease in expression of antioxidant genes affecting the NRF2-ARE pathway. Oxidative stress can also be increased by the stimulation of ROS production via increased NADPH oxidase activity or from disrupted mitchondrial respiratory chain activity. Mitochondrial activity can be affected by several ALS mutations, such as those leading to the accumulation of protein aggregates, or to decreased mitochondrial biogenesis and transport, or to increased cytosolic Ca2+ (as observed when glutamate receptor activity is stimulated or when the Ca2+-buffering capacity is decreased). Consequently a disruption of the mitochondrial respiratory chain will lead to an increase in ROS production and, thus, to an accumulation of oxidized proteins, lipids, DNA, and RNA. Oxidative damage occurring over time may then stimulate apoptotosis and, thus, cell death. Defective axonal transport affects not only the mitochondria but, also, the transport of other proteins and RNA, with consequences on the axon structure and function being accompanied by neurofilament accumulation. Defective glutamate uptake by astrocytes, and/or a defect in glutamate receptor clearance or in AMPA or GABA receptors, can lead to increased Ca2+ permeability and can impact the post-synaptic hyperexcitability and mitochondrial function. ARE: antioxidant response element, AMPA2: α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor 2, ATXN2: ataxin 2, Bcl2: B-cell lymphoma 2, C9orf72: Chromosome 9 open reading frame 72, C21orf2: Chromosome 21 open reading frame 7, CHCHD10: coiled-coil helix coiled-coil helix domain-containing 10, DCTN1: Dynactin 1, EEAT2: Excitatory amino acid transporter, ER: endoplasmic reticulum, FUS: Fused in Sarcoma, GABA: gamma-Aminobutyric acid, GlyR: glycine receptor, GlyT: glycine transporter, KIF5A: Kinesin heavy-chain isoform 5A, MAM: Mitochondria-associated ER membranes, NEFH: heavy-weighted neurofilaments, NEK1: (NIMA)-related kinase 1, NMDA: N-methyl-D-aspartate receptor, NRF2: Nuclear erythroid 2-Related Factor, PFN1: profilin-I, PTPIP51: Protein tyrosine phosphatase-interacting protein 51, SETX: senataxin, SOD: Superoxide dismutase 1, SPG11: Spatacsin, TDP-43: TAR DNA-binding protein 43, VAPB: vesicle-associated membrane protein-associated protein B, VCP: valosin-containing protein, and ROS: reactive oxygen species.