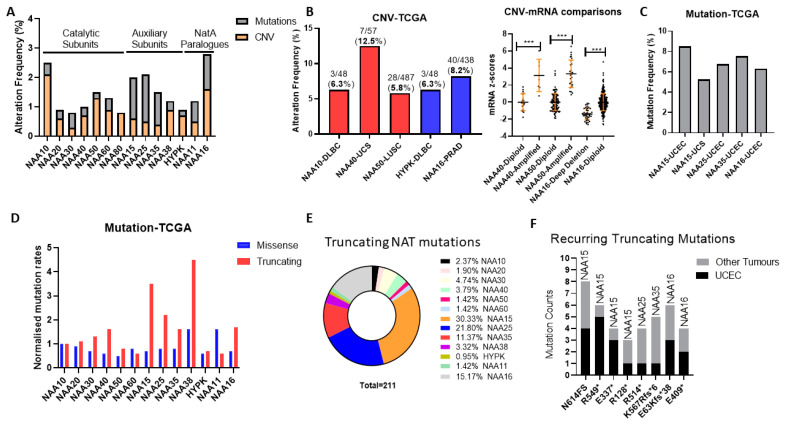
Figure 1.
Genomic alteration of N-terminal acetyltransferase (NAT) genes across The Cancer Genome Atlas (TCGA) pan-cancer study of 32 cancer types. (A) Oncoprint displaying average frequency of copy number variants (CNV) and mutations for each NATs across cancer types; (B) Tumour types with greater than 5% frequency of NAT amplifications or deletions (left panel) and association between amplification and mRNA levels in tumours (right panel). DLBC—Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, UCS—Uterine Carcinosarcoma, LUSC—Lung squamous cell carcinoma; (C) Tumour types with greater than 5% frequency of NAT mutations. UCEC—Uterine Corpus Endometrial Carcinoma; (D) counts of missense and truncating mutations for NAT genes across TCGA Pan-Cancer study normalised to protein length. Mutation counts for each NAT gene were normalised first to the length of the associated ORF and then to the corresponding value of NAA10. For NAA80, mutation data were not available; (E) Pie-chart depicting the frequency of truncating mutations for every NAT gene across the TCGA pan-cancer study; (F) Mutational count of recurring (at least three occurrences) truncating mutations of NAT genes in UCEC and other tumour types. *** p < 0.001 Student’s t-test.