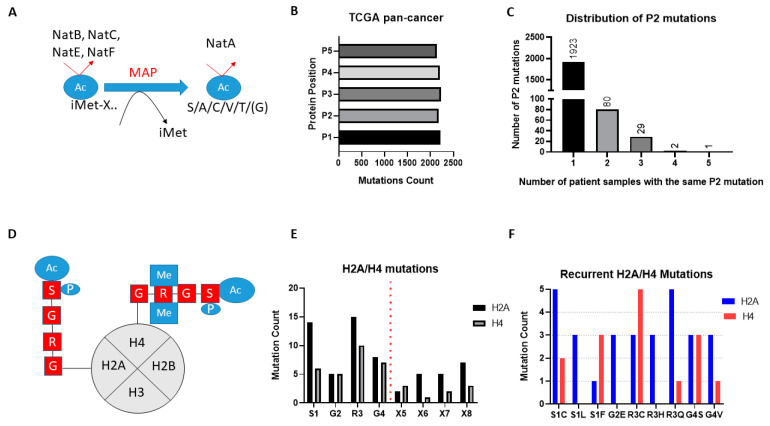
Figure 2.
Mutations affecting NAT protein substrates across 32 cancer types. (A) Schematic of Nt-Ac of broad-spectrum NATs. NatB, NatC, NatE, NatF directly methylate the iMET of substrate proteins. The substrate specificity of these NATs is determined by the amino acid adjacent to the methionine. After removal of iMET proteins through the action of MAP, N-termini with the first amino acid being S/A/C/T/V/G can be Nt-Ac through the action of NatA; (B) Missense mutational count for first five amino acids (P1–P5) across TCGA pan-cancer; (C) Distribution of missense mutations of P2 of proteins across TCGA pan-cancer; (D) Schematic of NAA40 recognition and Nt-Ac of histone H2A and H4; (E) Total missense mutational count of the first four amino acids (SRGR) across TCGA pan-cancer study; (F) Counts of recurring missense mutations (at least two occurrences) affecting the SRGR sequence in H2A or H4.