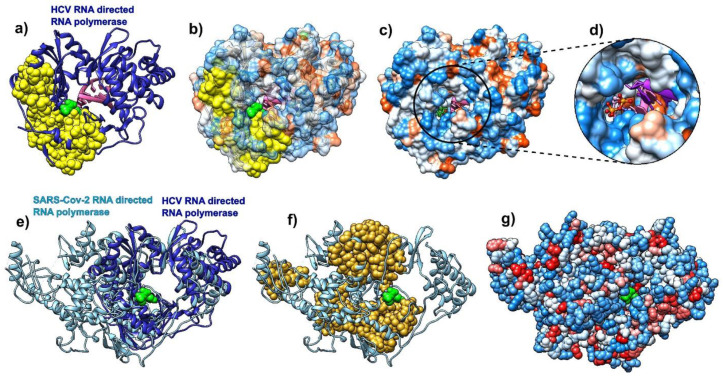
Figure 4.
Comparative analysis of hepatitis C virus (HCV) (pdb id 4wtg, chain A, left) bound to sofosbuvir, and COVID-19 RNA directed RNA polymerase (RdRp, pdb id 6m71, chain A, right). (a) HCV RNA-directed RNA polymerase is depicted as a blue ribbon, RNA is purple, and sofosbuvir is a green molecule (full atom representation). (b) HCV RdRp is represented as a transparent hydrophobic surface, SAGNM predictions are yellow and sofosbuvir is represented via green atoms. (c) HCV RdRp is represented as an opaque hydrophobic surface, and sofosbuvir is represented via green sticks. (d) The inset shows sofosbuvir inside the polymerase catalytic core. (e) HCV RdRp (blue ribbon) structurally aligned with COVID-19 RdRp (light blue ribbon). Sofosbuvir is a green molecule inside the HCV RdRp catalytic core. (f) COVID-19 RdRp as a light blue ribbon. SAGNM predictions are dark yellow atoms. Sofosbuvir is a green molecule inside the catalytic core. The position stems from the structurally aligned HCV RdRp. (g) COVID-19 RdRp as hydrophobically colored atoms (residues hydrophobicities). Sofosbuvir is a green molecule inside the catalytic core. The position stems from the structurally aligned HCV RdRp. With COVID-19 RNA polymerase, sofosbuvir’s position corresponds to the position it has when bound to HCV RNA polymerase.