Figure 2. MAS-DNP methods for probing protein-ligand binding.
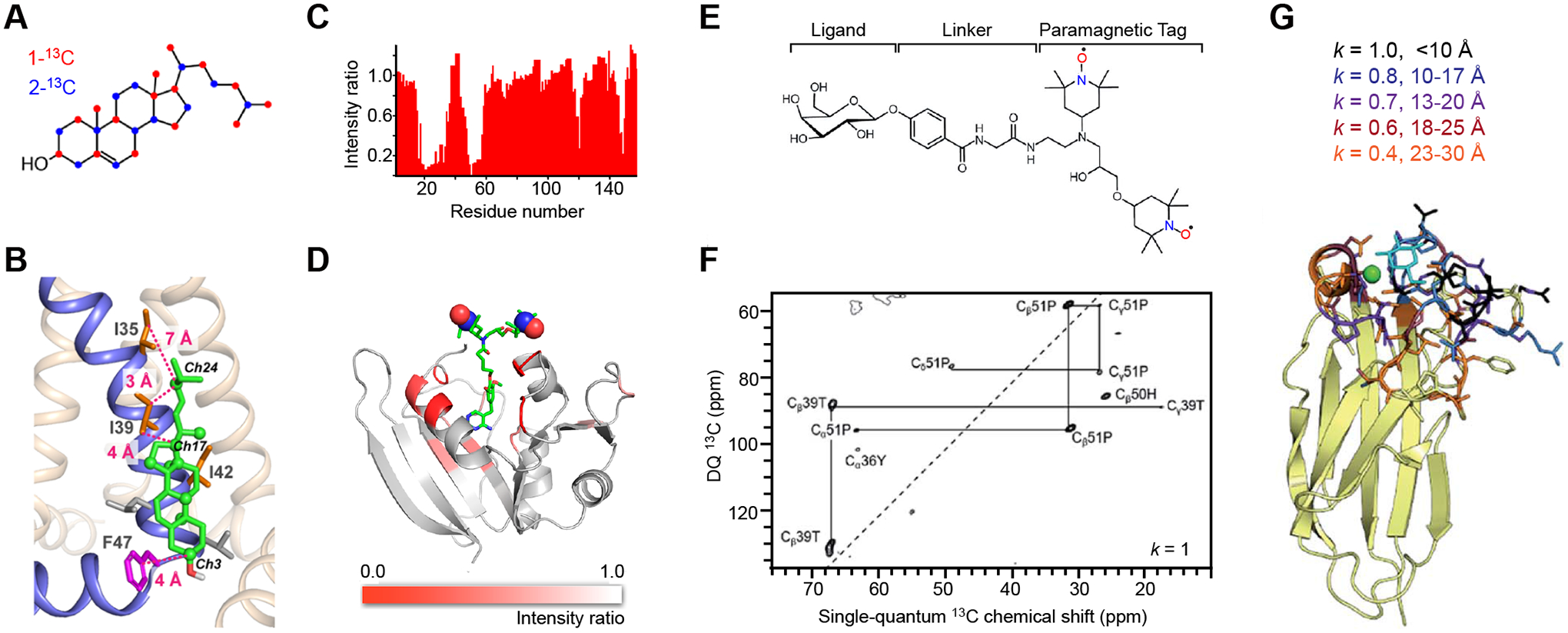
(A) Yeast-based 13C labeling of cholesterol using site-specifically 13C-labeled glucose. The labeled carbon sites on cholesterol are in red and blue for cholesterols produced from 1-13C and 2-13C glucose molecules, respectively. (B) A structural model of a cholesterol molecule bound to the influenza M2 proteins. The key Ile and Phe residues, as well as their distances to cholesterol carbons, are shown. (C) Signal bleaching quantified in solution 1H-15N HSQC spectra due to the binding of radicals to dihydrofolate reductase. (D) A model of E. coli dihydrofolate reductase with DNP bleaching information represented by the intensity ratios of 13C-13C DARR spectra collected on two samples containing either bound radicals or exogenous radicals. (E) Scheme for incorporating a carbohydrate ligand to a paramagnetic tag for selective DNP. (F) Selective DNP 13C–13C INADEQUATE difference spectrum of LecA obtained using k=1: only the tightly bound residues are observed. (G) Sideview of LecA. Residues observed using selective DNP are highlighted, with the corresponding k values given. Figures 2A-2D are adapted from references [38, 51] with copyright permission. Figures 2E-2G are adapted from reference [31], an open-access article.
