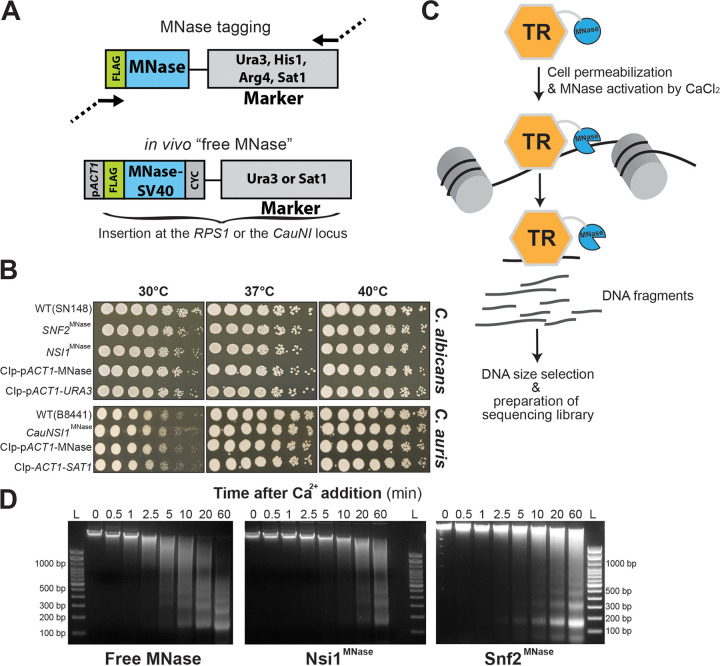
FIG 1.
ChEC-seq method in C. albicans and Candida spp. (A) Plasmid constructs for in vivo TR-MNase tagging and the construction of the “free MNase” control strains in C. albicans and C. auris. Structure of the MNase-tagging cassette consisting of a 3xFLAG epitope fused to the C. albicans codon-optimized MNase. The FLAG-MNase construct for the “free MNase” strain was constitutively expressed using the C. albicans ACT1 promoter (pACT1). CYC, CYC1 terminator; SV40, nuclear localization signal. (B) Phenotypic characterization of strains bearing the MNase-tagged Nsi1 and Snf2 TRs and the “free MNase” control constructs in C. albicans. WT, Snf2MNase, Nsi1MNase, the free MNase, and the control (empty vector) strains were serially diluted, spotted on YPD, and incubated for 1 day at different temperatures. Growth patterns at different temperatures of the C. auris WT, CauNsi1MNase, the free MNase and the control strains are also shown. (C) Schematic representation of the experimental setup of the ChEC-seq methodology. Candida species cells where a TR of interest is fused to MNase are permeabilized with digitonin prior to MNase activation with calcium. This will lead to the fragmentation of unprotected neighboring chromatin. The resulting fragmented DNA is purified and subjected to size selection prior to high-throughput sequencing. (D) Evaluation of genomic DNA fragmentation by agarose gel electrophoresis at 0, 0.5, 1, 2.5, 5, 10, 20, and 60 min of calcium exposure in the Snf2MNase, Nsi1MNase, and the free MNase strains in C. albicans. L, 100-bp DNA ladder.