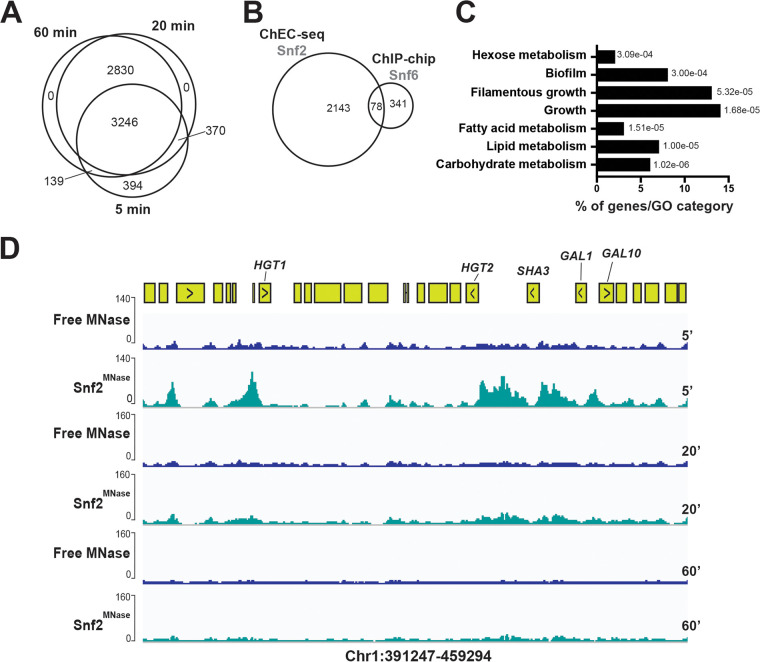
FIG 3.
Genome-wide occupancy of the chromatin remodeler Snf2 with ChEC-seq. (A) Venn diagram showing the overlap of Snf2 promoter binding at 5, 20, and 60 min after MNase activation. (B) Comparison of SWI/SNF genomic occupancies by ChEC-seq and ChIP-chip method. Venn diagram of overlap between Snf2 ChEC-seq sites and Snf6 ChIP-chip using high-density tiling arrays. (C) Gene ontology of biological process associated with Snf2-bound promoters at 5 min after MNase activation. The P values were calculated using hypergeometric distribution as described in the GO Term Finder Tool website (http://candidagenome.org/cgi-bin/GO/goTermFinder). (D) Snapshot of genomic regions showing the ChEC-seq signal for Snf2MNase and the free MNase strains at 5, 20, and 60 min after MNase activation. Genome browser view of Snf2 ChEC-seq displaying promoter occupancies of carbohydrate metabolism genes, including galactolysis (GAL1 and GAL10) and hexose transport (SHA3, HGT2, and HGT1).