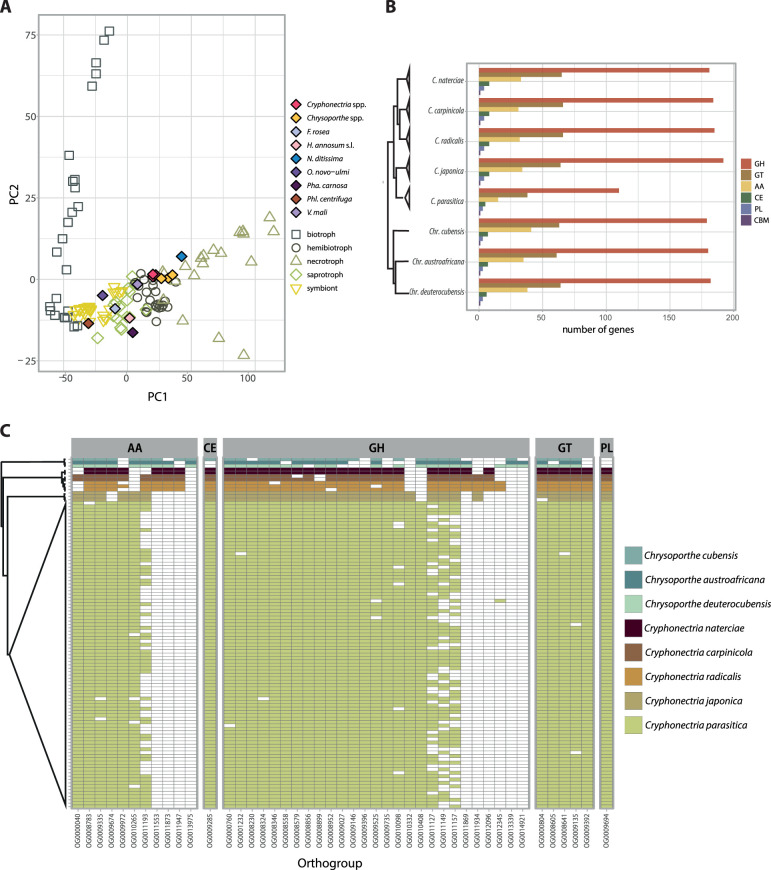
FIG 2.
Carbohydrate-active enzyme (CAZyme) content among Cryphonectriaceae. (A) Principal-component analysis (PCA) of fungal lifestyle predictions, as inferred by CATAStrophy. The plot incorporates 85 reference species of fungi with different lifestyles (i.e., biotroph, hemibiotroph, nectrotroph, saprotroph, and symbiont) used as a training set by CATAStrophy and shows the CAZyme-inferred phenotypic trophism of Cryphonectriaceae and other pathogenic and nonpathogenic tree-associated fungi. (B) Number of detected CAZyme genes per species grouped according to CAZyme superfamily: glycoside hydrolase (GH), glycosyl transferase (GT), auxiliary activity (AA), carbohydrate esterase (CE), polysaccharide lyase activity (PL), and carbohydrate-binding modules (CBM). (C) Ortholog presence/absence of CAZyme superfamilies for which at least one species is missing an ortholog (the CBM superfamily is not shown, as orthologs were found in all species).