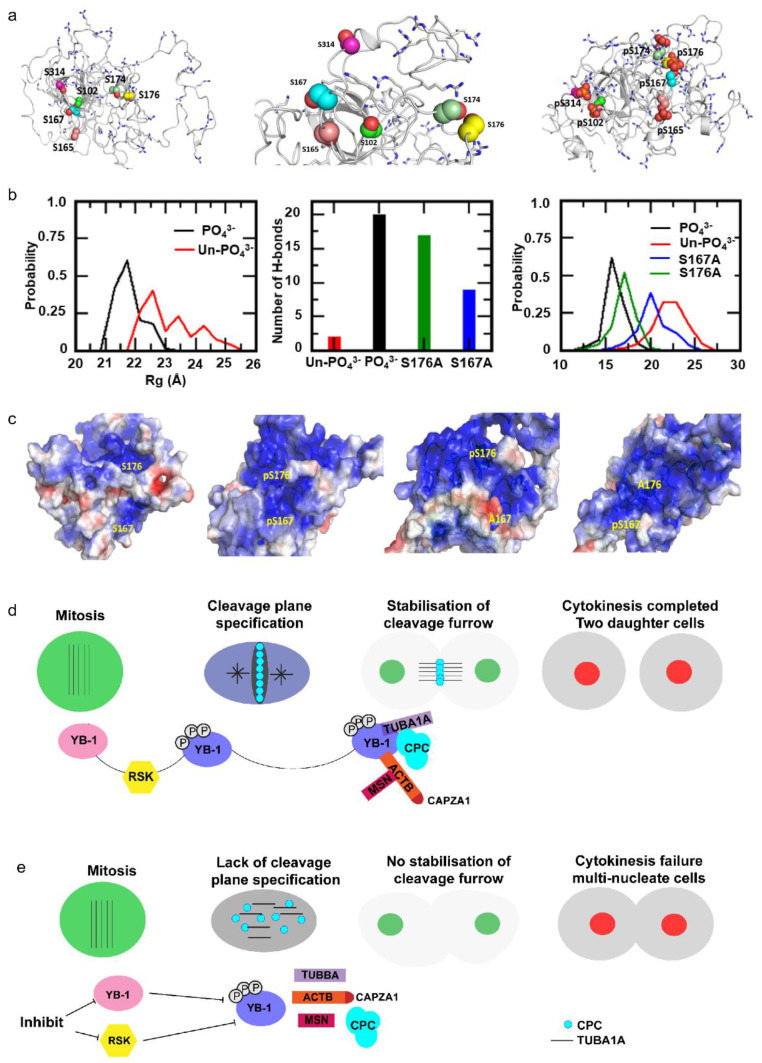
Figure 8.
Molecular dynamics simulation models the impact of phosphorylation on localization of the CPC to the cleavage furrow. (a) YB-1 model generated using I-TASSER and MD simulations, left: un-phosphorylated, middle: zoomed view of serines and right: phosphorylated states (pYB-1). Spheres represent serines and phosphoserines, thick grey lines represent positively-charged Arg and Lys residues. The ordered cold shock domain is shown at the centre flanked by the disordered long termini. (b) Left: Distribution of the radius of gyration (Rg), a measure of protein compaction. Middle: number of hydrogen bonds involving the serines/phosphorylated serines. Right: distribution of the distances between the backbone atoms of residues 167 and 176 sampled during the MD simulations of YB-1 and phosphorylated YB-1. (c) Electrostatic surfaces projected on to (left to right) YB1 (unphosphorylated), pYB-1 (phosphorylated), and the two mutants S167A and S176A; the blue and red colours correspond to potentials of +3 kcal/mol and −3 kcal/mol, respectively. (d) YB-1 has a disordered structure with neutral and negatively charged pockets. Upon phosphorylation by RSK, YB-1 protein undergoes a structural change, creating a positively charged pocket. We speculate that this pocket interacts with the negatively charged termini of α-tubulin to define the cleavage plane by facilitating the formation of the microtubule structure and localization of the CPC, which, in turn, leads to the stabilization of the cleavage furrow. YB-1 also interacts with β-actin and actin related proteins (CAPZA1 and MSN), promoting cytokinesis. (e) When phosphorylation of YB-1 is inhibited or depleted, the cleavage plane defined by the α-tubulin scaffold cannot form resulting in a failure of CPC localization and cytokinesis.