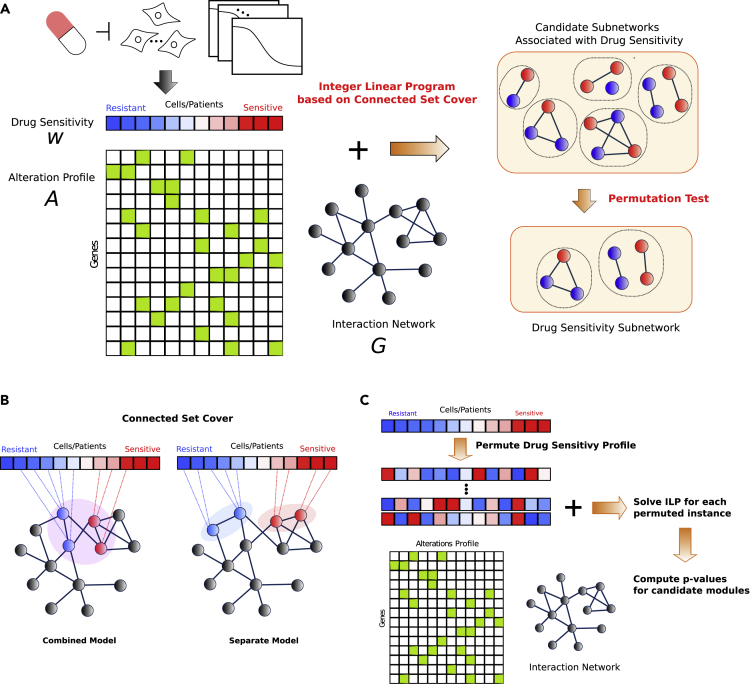
Figure 1.
NETPHIX Method
(A) Overview: NETPHIX takes drug sensitivity profile, alterations status for the same set of samples, and interaction information among genes as inputs. Using a connected set cover-based ILP algorithm, we first generate a set of candidate modules. The final set of modules include only maximal modules among statistically significant solutions. Genes associated with decreased and increased sensitivity are marked as blue and red, respectively.
(B) NETPHIX finds a connected set of genes of which alterations are associated with phenotype values (red colors in the drug response profile indicate increased sensitivity values and blue colors are for decreased sensitivity values). We considered the combined model in which all the selected genes are connected and the separate model in which two subnetworks are identified for increased and decreased sensitivity separately.
(C) The significance of identified modules is assessed using a permutation test by permuting drug sensitivity profiles.