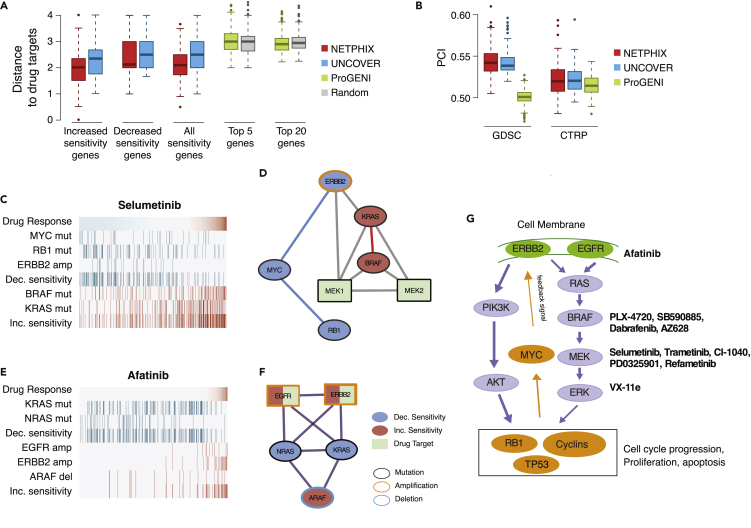
Figure 3.
Evaluation Results and Modules Identified with Real Drug Screening Dataset
(A–B) Evaluation with drug screening dataset. (A) Average distances of selected genes to drug targets. Distances for genes associated with increased sensitivity, decreased sensitivity, all genes in the selected modules are shown for NETPHIX and UNCOVER. The distances with 5 and 20 top ranked genes for ProGENI and randomly selected genes are also shown. (B) Predictive power of the modules selected by NETPHIX, UNCOVER, and ProGENI. PCI scores with random forest regression tested within GDSC and with CTRP dataset are shown.
(C–F) Sensitivity networks identified by NETPHIX. Alternation profile and connectivity between the drug target and the genes for Selumetinib (C, D) and Afatinib (E, F). In the alteration profile, the panel shows the values of the phenotype (i.e., drug response, top row) for all samples (columns), with blue being decreased sensitivity values and red being increased sensitivity values. For each gene, alteration status in each sample is shown in red/blue (with the summary covers for decreased and increased sensitivity separately), whereas samples not altered are shown in gray. The module for Selumetinib is identified based on the separate connectivity model, and the module for Afatinib is selected based on the combined connectivity model.
(G) Schematic diagram of MAPK/ERK and AKT signaling pathways with drugs and their drug targets annotated.