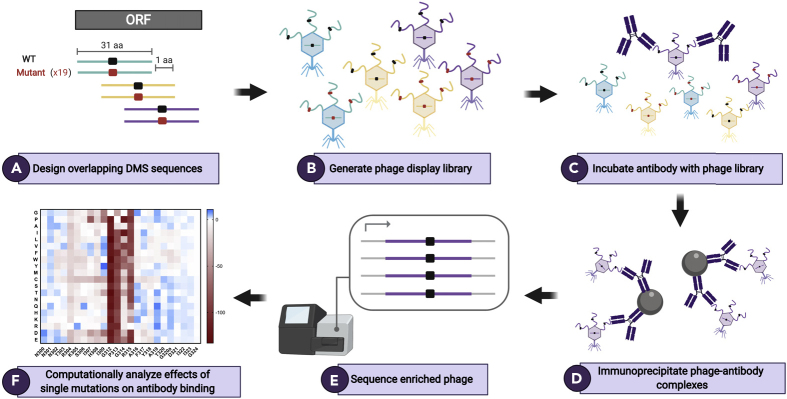
Figure 1.
Schematic of Phage-DMS Library Design and Experimental Approach
(A) To build a Phage-DMS library, sequences are computationally designed to tile across an entire open reading frame of interest, with the central position varying to contain either the wild-type residue (shown in black) or a mutant residue (shown in red).
(B) Sequences are synthesized by releasable DNA microarray and then cloned into a T7 phage display vector.
(C and D) (C) The resulting phage display library is incubated with antibody, and (D) phage-antibody complexes are immunoprecipitated with magnetic beads.
(E) Sequences from enriched phage are PCR amplified and pooled samples are deeply sequenced.
(F) Finally, computational analysis is performed to determine the relative effect of single mutations on the binding of antibody to antigen. Figure made using BioRender.com.