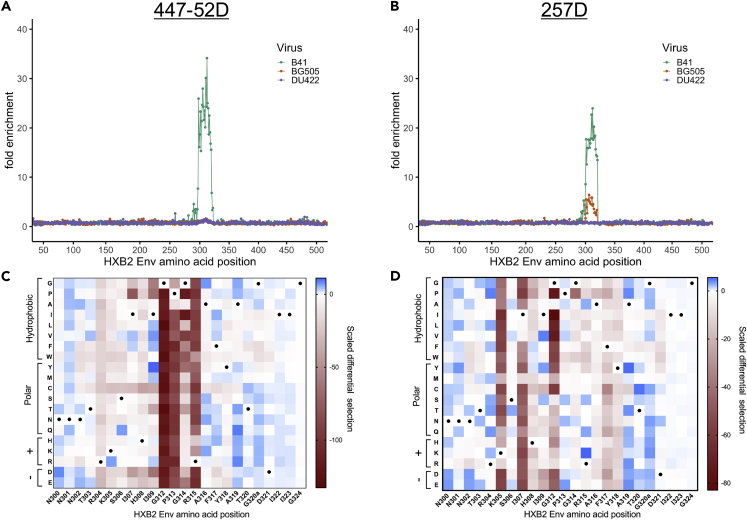
Figure 4.
Enrichment and Scaled Differential Selection Results from Phage-DMS for V3-Specific mAbs with gp120 Libraries
(A and B) Line plot showing fold enrichment of wild-type peptides in the background of each HIV Env strain for (A) mAb 447-52D and (B) mAb 257D.
(C and D) The color corresponding to each wild-type sequence is shown in the upper right. The x axis shows the amino acid position within HIV Env based on HXB2 reference sequence. (C and D) Heatmap showing the relative effect, as compared with wild-type B41 Env, of each mutation on the binding to (C) mAb 447-52D and (D) mAb 257D, within a selected region of V3 shown with HXB2 numbering. Wild-type residues are marked by a black dot. Amino acids are grouped based on their properties as indicated to the left. All data shown are the average of two biological replicates.
See Quantification and Statistical Analysis for fold enrichment and scaled differential selection calculations.