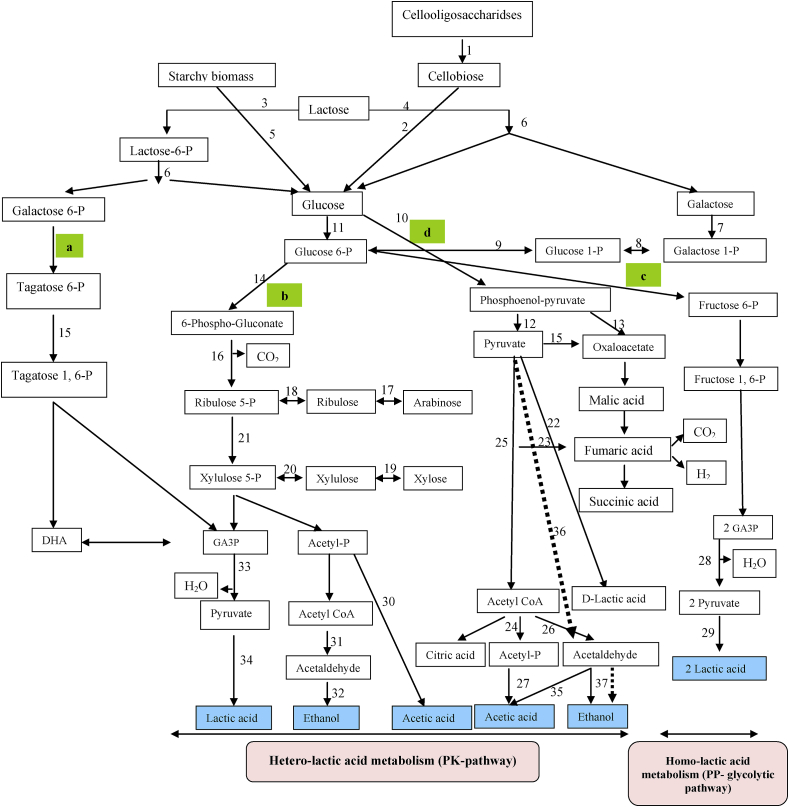
Figure 1.
Pathways of lactic acid production from agro-industrial residues. Number on arrow catalyzed by enzyme and other reaction. 1: Exo β1,4 Glucanase, 2: β -Glucosidase, 3: lactose phosphotransferase system (Lac-PTS), 4: permease, 5: Amylase, 6: β-galactosidase, 7: ATP→ADP, 8: galactose-1-phosphate uridylyltransferase, 9: phosphoglucomutase, 10: NAD→ NADH, 11: ATP→ADP, 12: ATP→ADP, 13: Phosphoenolpyruvate carboxylase, 14: ATP→ADP, 15: ATP→ADP, 16: NAD+→NADH, 17: arabinose isomerase, 18: ribulokinase and ATP→ADP, 19: xylose reductase and xylitol dehydrogenase, 20: ATP→ADP, 21: ribulose 5-phosphate 3-epimerase, 22: D-lactic acid Dehydrogenase, 23: Pyruvate-fomarate lyase, 24: Pta, 25: Pyruvate dehydrogenase complex, 26: Aldehyde dehydrogenase, 2NADH→ 2NAD+, 27: Acetate kinase, 28: 4 ADP→ 4ATP, 2 NAD+→2NADH, 29: 2NADH→2NAD+, 30: ADP→ ATP, 31: NADH→ NAD+, 32: NADH→ NAD+, 33: 2ADP→ATP, NAD+→NADH, 34: Lactate dehydrogenase, NADH→NAD+, 35: Acetaldehyde dehydrogenase, 36: Pyruvate decarboxylase. 37: Alcohol dehydrogenase. GA3P: glyceraldehyde-3-P, DHAP: Dihydroxyacetone-P. A route: D-tagatose 6-phosphate pathway. B route: Pentose phosphoketolase (PK) pathway: for Hetero lactic acid metabolism. C route: Embden-Meyerhof-Parnas (EMP) pathway: for Homo lactic acid metabolism. D route: Glycolysis pathway in E. coli, K. lactis and S. cerevisiae.