Fig. 1. Structure determination: transcription-translation complexes (TTCs).
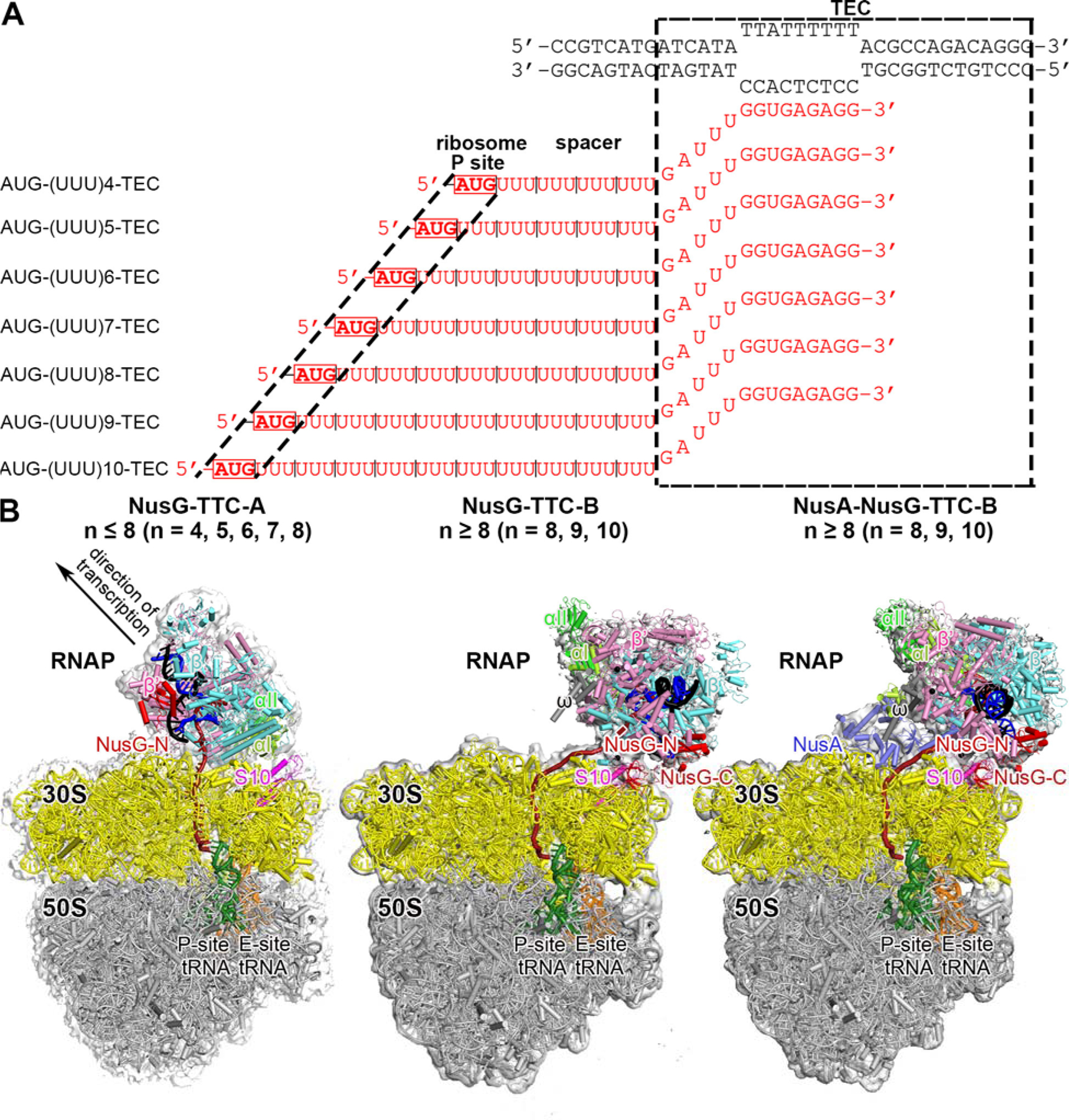
(A) Nucleic-acid scaffolds. Each scaffold comprises nontemplate- and template-strand oligodeoxyribonucleotides (black) and one of seven oligoribonucleotides having spacer length n of 4, 5, 6, 7, 8, 9, or 10 codons (red), corresponding to mRNA. Dashed black box labeled “TEC,” portion of nucleic-acid scaffold that forms TEC upon addition of RNAP (10 nt nontemplate- and template-strand ssDNA segments forming “transcription bubble,” 10 nt of mRNA engaged with template-strand DNA as RNA-DNA “hybrid,” and 5 nt of mRNA, on diagonal, in RNAP RNA-exit channel); dashed black lines labeled “ribosome P-site,” mRNA AUG codon intended to occupy ribosome active-center P site upon addition of ribosome and tRNAfMet; “spacer,” mRNA spacer between TEC and AUG codon in ribosome active-center P site.
(B) Cryo-EM structures of NusG-TTC-A (obtained with spacer lengths of 4–8 codons), NusG-TTC-B (obtained with spacer lengths of 8–10 codons), and NusA-NusG-TTC-B (obtained with spacer lengths of 8–10 codons). Structures shown are NusG-TTC-A (3.7 Å; n = 4; Table S1), NusG-TTC-B (4.7 Å; n = 9; Table S1), and NusA-NusG-TTC-B2 (3.5 Å; n = 8; Table S1). Images show EM density (gray surface) and fit (ribbons) for TEC, NusG and NusA (at top; direction of transcription, defined by downstream dsDNA, indicated by arrow in left panel and directly toward viewer in center and right panels) and for ribosome 30S and 50S subunits and P- and E-site tRNAs (at bottom). RNAP β’, β, αI, αII, and ω subunits are in pink, cyan, light green, and dark green, and gray; 30S subunit, 50S subunit, P-site tRNA, E-site tRNA are in yellow, gray, green, and orange; DNA nontemplate strand, DNA template strand, and mRNA are in black, blue, and brick-red (brick-red dashed line where modelled). NusG, NusA, and ribosomal protein S10 are in red, light blue, and magenta. Ribosome L7/L12 stalk omitted for clarity in this and all subsequent images.
