FIGURE 5.
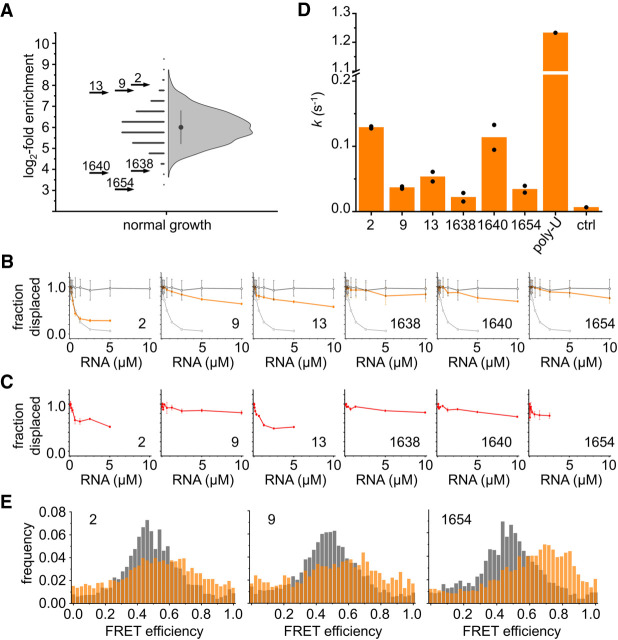
Binding of selected RNAs to Hera in vitro. (A) Half-Violin plot showing the range of log2-fold enrichment for all RNA sequences identified under normal-growth conditions. On the left side, the log2-fold change for each RNA is indicated by a symbol, the right-hand side shows the distribution. The filled circle marks the mean log2-fold enrichment, the line indicates one standard deviation. The log2-fold enrichment of the selected RNAs 2, 9, and 13 (log2-fold enrichment 8.0, 7.7, and 7.6) and 1638, 1640, and 1654 (log2-fold enrichment 3.9, 3.9, and 3.0) is indicated by arrows. (B) Displacement titrations of a Hera/32mer complex with RNAs 2, 9, 13, 1638, 1640, and 1654. The fluorescence anisotropy of a fluorescein attached to the 5′-end of the 32mer was used as a probe for 32mer dissociation due to binding of unlabeled RNAs. The open black circles depict values for a control experiment with a nonbinding 50A-RNA (negative control), the open gray circles show the control experiment with a binding 50U-RNA (positive control; see Supplemental Fig. 6). The average values from two replicate experiments are shown; error bars denote the standard error of the mean (SEM). Values are normalized: A value of one corresponds to no displacement of the fluorescein-labeled 32mer, a value of zero corresponds to complete displacement. (C) Displacement titrations of an RBD/32mer complex with RNAs 2, 9, 13, 1638, 1640, and 1654. The average of two replicate experiments is shown. Values are normalized: A value of one corresponds to no displacement of the fluorescein-labeled 32mer, a value of zero corresponds to complete displacement. (D) Rate constants k for ATP hydrolysis by Hera in the presence of different RNAs (10 µM) and saturating concentrations of poly(U) RNA (500 µM bases, see Linden et al. 2008) as an internal control. Experiments were performed in duplicate; individual values are depicted as circles, the bars show the mean. Ctrl: background in the presence of poly(U) and absence of enzyme. (E) Normalized single-molecule FRET histograms of Hera without RNA (gray), and in the presence of RNA and nucleotide (orange). RNA 2 and to a lesser extent also RNA 9 causes a shift in FRET efficiency to higher values, indicating that the Hera core closes in part of the Hera molecules. RNA 1654 induces a clear shift in FRET efficiency to higher values, consistent with closure and activation of the helicase core (see Supplemental Fig. 9 for control experiments).