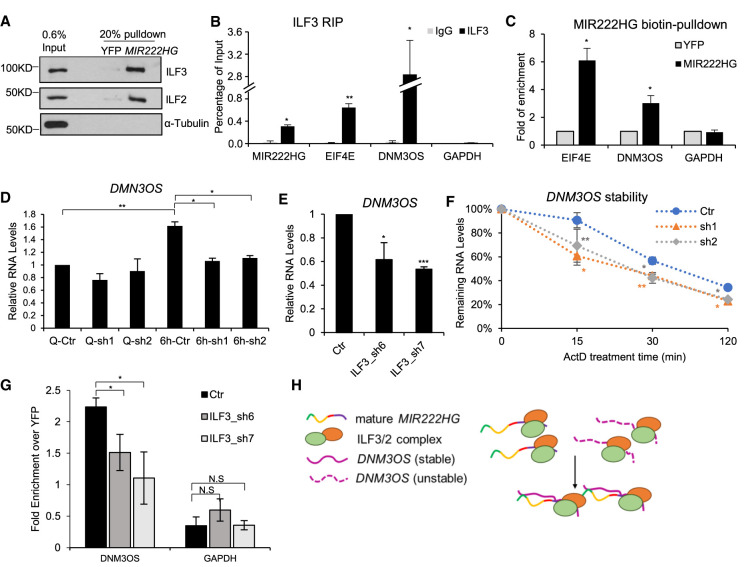
FIGURE 6.
MIR222HG forms RNP complex containing ILF3/2 protein and other RNAs. (A) In vitro biotin-RNA pull-down followed by immunoblotting to detect the interaction between MIR222HG and ILF3/ILF2. YFP RNA is used as negative RNA control. α-Tubulin is used as negative protein control. (B) ILF3 RIP followed by RT-qPCR to quantify the interaction between ILF3 and MIR222HG or EIF4E (3′-UTR region) or DNM3OS lncRNA in WI-38 cells. GAPDH is used as negative control. (C) In vitro biotin-RNA pull-down followed by RT-qPCR to determine the interaction between MIR222HG and ILF3/2-target RNAs in WI-38 cells. Relative fold enrichment of EIF4E mRNA and DNM3OS lncRNA are calculated by comparing to biotin-YFP. GAPDH is used as negative control. (D) RT-qPCR to quantify the levels of DNM3OS lncRNA in control and MIR222HG-depleted WI-38 cells in quiescent and 6-h post-serum-stimulated WI-38 cells. (E) RT-qPCR to quantify the levels of DNM3OS in control and ILF3-depleted WI-38 cells post 6-h serum-stimulation. (F) RNA stability analyses of DNM3OS in control and MIR222HG-depleted WI-38 cells. RNA is extracted from cells incubated with actinomycin D for indicated time points (min). Relative levels of RNA are measured via RT-qPCR. (G) In vitro biotin-RNA pull-down followed by RT-qPCR to determine the interaction between MIR222HG and DNM3OS in control and ILF3-depleted serum-stimulated WI-38 cells. Relative fold enrichment of DNM3OS lncRNA is calculated by comparing to biotin-YFP. GAPDH is used as negative control. (H) Schematic of ILF3/2-MIR222HG-DNM3OS RNP complex. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001 by two-tailed Student's t-test, n = 3 for all figures. Error bars represent standard deviation.