FIGURE 5.
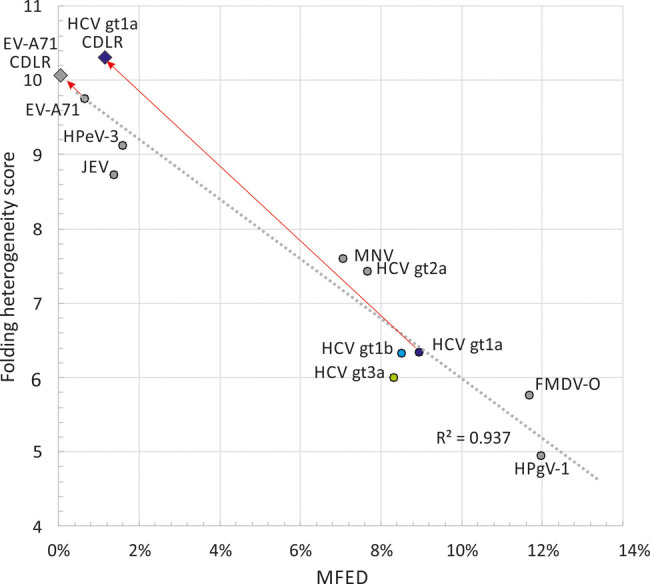
Association of MFED scores with folding prediction heterogeneity. Comparison of MFED scores with folding prediction heterogeneity for complete polyprotein sequences of HCV, other predicted structured viruses (HPgV-1, FMDV-O, and MNV), and unstructured viruses (JEV, HPeV-3, and EV-A71). Folding heterogeneity was expressed as the difference in folding depth between different sequences within an alignment averaged over the whole polyprotein sequence. As controls, sequence order randomized sequence data sets were generated by the algorithm CDLR for structured (HCV-1a) and unstructured (EV-A71) genome sequences and their folding energies (MFED score; x-axis) and structural heterogeneity (y-axis) reestimated.
