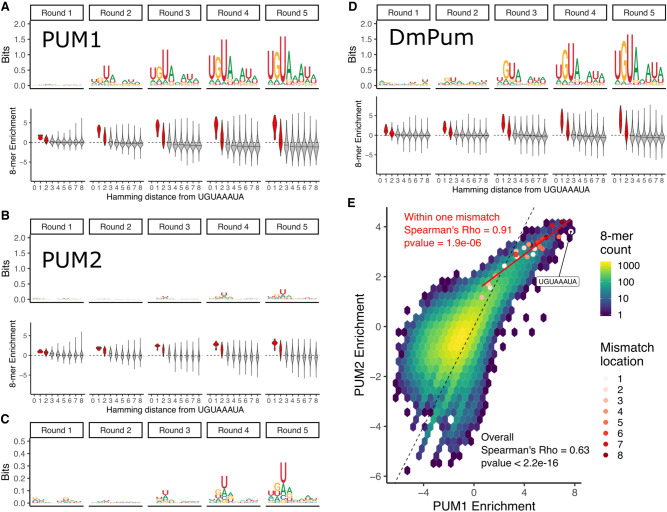
FIGURE 2.
SEQRS analysis of Human PUM1 and PUM2 PUM-HDs reveals preference for the canonical PUM recognition element. (A, top) Position weight matrices representing 8-mer sequence preferences for purified Human PUM1 PUM-HD, as determined for each SEQRS round. (Bottom) 8-mer enrichment, as measured by log2(Enrichment SEQRS round/Enrichment no protein) (see Materials and Methods for details) for each 8-mer as binned by Hamming distance from the canonical UGUAAAUA PUM recognition element. Enrichment scores for 8-mers within two mismatches are filled in red. (B) Same as in A, but for Human PUM2 PUM-HD. (C) Closer view of Human PUM2 PUM-HD PWMs. (D) Same as in A, but for Drosophila Pum PUM-HD. (E) Correlation of 8-mer enrichment between Human PUM1 and Human PUM2 PUM-HDs. Enrichment for all possible 8-mers are displayed in a two-dimensional histogram. The dashed black line represents one-to-one correspondence. All 8-mers within one mismatch to the UGUAAAUA sequence are plotted as red points with the color specifying the position within the motif where the mismatch occurs. The red line is a linear fit using only the UGUAAAUA 8-mer and all 8-mers within one mismatch.