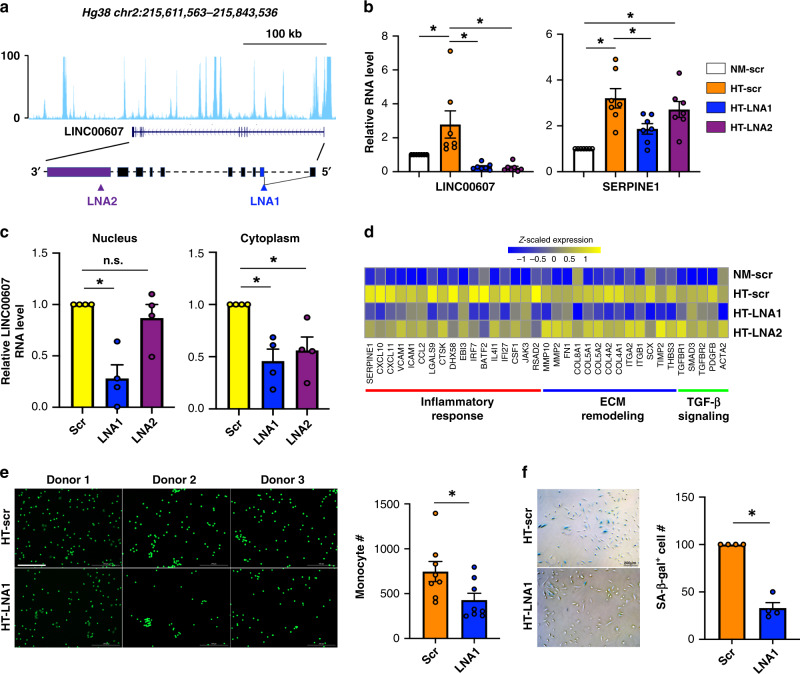
Fig. 4. Inhibition of LINC00607 attenuates SERPINE1 induction and EC dysfunction.
a Illustration of LINC00607 genomic locus and gene structure and LNA GapmeRs targeting of LINC00607 RNA. b HUVECs were transfected with scramble (scr) or two LNAs targeting LINC00607. LINC00607 RNA and SERPINE1 mRNA levels were quantified. Data represent mean ± SEM from seven independent experiments. c qPCR of LINC00607 in subcellular fractionations of HUVECs transfected with scr, LNA1, or LNA2. Data represent mean ± SEM from four independent experiments. d RNA-seq was performed with cells transfected as in b in biological replicates. Heatmap is plotted based on z-scaled log-transformed gene expression levels. e, f ECs transfected with scr or LNA1 and then treated by H + T were used in e monocyte adhesion assay and in f SA-β-gal assay. In e, the number of attached monocytes to ECs were quantified. Data represent mean ± SEM from eight independent experiments performed with peripheral blood-derived monocytes from four individual donors. Representative images show the attachment of fluorescently labeled peripheral blood-derived monocytes to ECs for experiments performed with monocytes from three different donors. In f, ECs with positive β-gal staining were quantified. The positively stained cell number in scr control was set to 100 (%). f Data represent mean ± SEM from four independent experiments respectively. Scale bar = 200 μm. * denotes p = 0.0289, 0.0013, 0.001, 0.0001, 0.0025, and 0.024 from left to right (in b) and p = 0.0023, 0.0079, and 0.0249 from left to right (in c) between indicated groups based on ANOVA followed by Bonferroni post hoc test (in b and c); p = 0.0039 (in e) and 0.0014 (in f), as compared to scr group based on two-tailed paired t test (in e and f). Source data are provided as a Source data file.