Figure 2.
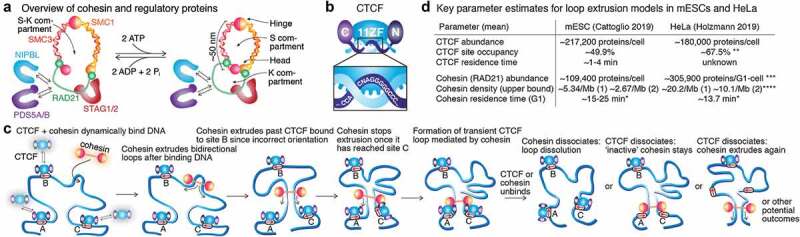
Overview of cohesin, CTCF, and loop extrusion. (a) Overview of mammalian cohesin and some of its regulatory proteins. (b) Overview of CTCF with N-terminal, 11 Zinc Fingers, and C-terminal domains. (c) Simplified sketch of cohesin-mediated loop extrusion and the convergent CTCF rule. (d) Summary of key parameters constraining loop extrusion models in mouse embryonic stem cells (mESCs) [65] and human HeLa cells [83], with mESC residence times taken from [70]. * These are cohesin G1 residence times (both STAG1 and STAG2), but after these studies were published it was found that STAG2-cohesin binds DNA substantially more dynamically than STAG1-cohesin [38], suggesting that putative loop extruding G1 cohesins have at least two residence times. ** Estimated from [83] (~180,000 and ~120,000 CTCF proteins and sites per HeLa cell) with added assumption that 45% of CTCF proteins are bound to cognate sites (~45%, i.e. mean of mESC and U2OS in [70]). *** 305,900 is the mean of the LC-MS and FCS estimates reported in [83]. **** Cohesin density is estimated from ~159,437 dynamically bound (~13.7 min residence time) cohesin proteins (SCC1-mEGFP) in G1 and the reported HeLa genome sizes 7.9 Gb, both taken from [83]. It is important to note that these are genomic averages: e.g. CTCF residence time is for an average site (some sites will have slower and faster binding), cohesin density may not be uniform throughout the genome, and since the two in vitro cohesin loop extrusion papers disagreed on whether cohesin is monomeric [23] or dimeric [21], densities for both monomeric [1] and dimeric [2] are shown.
