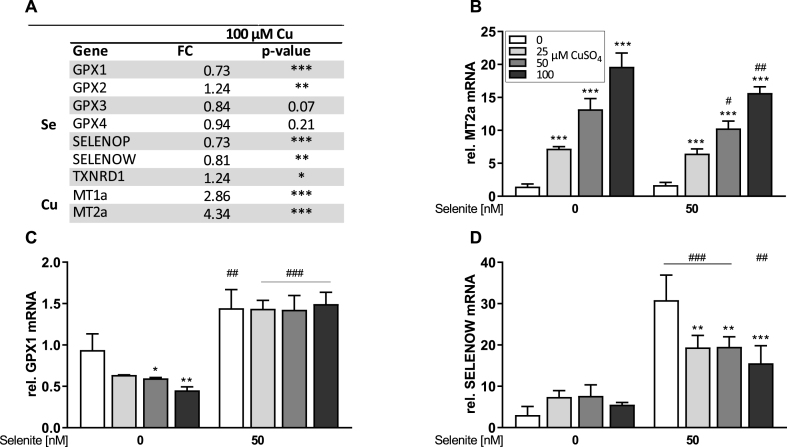
Fig. 1.
Expression of Se- and Cu-dependent genes in HepG2 cells. Microarray data provided by GEO Profiles (GEO Series Accession No. GSE9539) [35] obtained from HepG2 cells treated for 24 h with 100 μM CuSO4 (A). Data are given as fold change (FC) relative to the untreated control (n = 3). qPCR results of various Se- and Cu-responsive genes analyzed in HepG2 cells cultured with increasing Cu concentrations (0, 25, 50 or 100 μM) combined with or without 50 nM selenite for 48 h (B–D). Gene expression was normalized to the reference genes RPL13A and HPRT. Untreated cells of the first replicate were set as 1. Data are depicted as mean + SD (n = 3). Statistical analyses were based on two-way ANOVA with Bonferroni's post-test. *p < 0.05; **p < 0.01; ***p < 0.001 vs. 0 μM CuSO4 and #p < 0.05; ##p < 0.01; ###p < 0.001 vs. 0 nM Se.