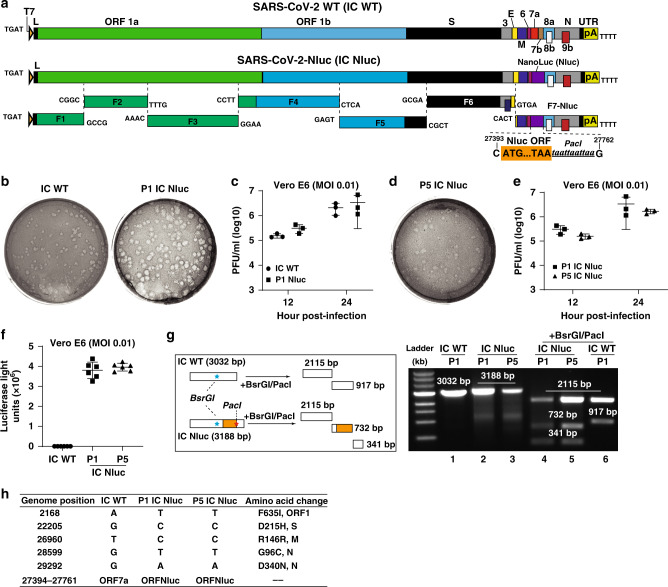
Fig. 1. Development and characterization of SARS-CoV-2-Nluc.
a Assembly of the full-length SARS-CoV-2-Nluc cDNA. The Nanoluciferase (Nluc) gene together with a PacI site was placed downstream of the regulatory sequence of ORF7 to replace the ORF7 sequence. The nucleotide identities of the Nluc substitution sites are indicated. b Plaque morphologies of infectious clone-derived P1 SARS-CoV-2-Nluc (P1 IC Nluc) and wild-type SARS-CoV-2 (IC WT). c Replication kinetics. Vero E6 cells were infected with infectious clone-derived IC WT or P1 IC Nluc at MOI 0.01. Viruses in culture supernatants were quantified by plaque assay. The means ± standard deviations from three independent experiments are shown. Two-way ANOVA with correct for multiple comparisons are used for statistical analyses. d Plaque morphology of P5 IC Nluc. e Replication kinetics of P5 IC Nluc on Vero E6 cells. The means ± standard deviations from three independent experiments are shown. Two-way ANOVA with correct for multiple comparisons are used for statistical analyses. f Luciferase signals produced from SARS-CoV-2-Nluc-infected Vero E6 cells at 12 h post-infection. Cells were infected with viruses at MOI 0.1. The means ± standard deviations from six independent experiments are shown. One-way ordinary ANOVA test was used for statistical analyses. g Gel analysis of IC Nluc virus stability. The left panel depicts the theoretical results of RT-PCR followed by restriction enzyme digestion. The right panel shows the gel analysis of the RT-PCR products before (lanes 1–3) and after BsrGI/PacI digestion (lanes 4–6). h Summary of full-genome sequences of P1 and P5 IC Nluc viruses. Nucleotide and amino acid differences from the IC WT are indicated.