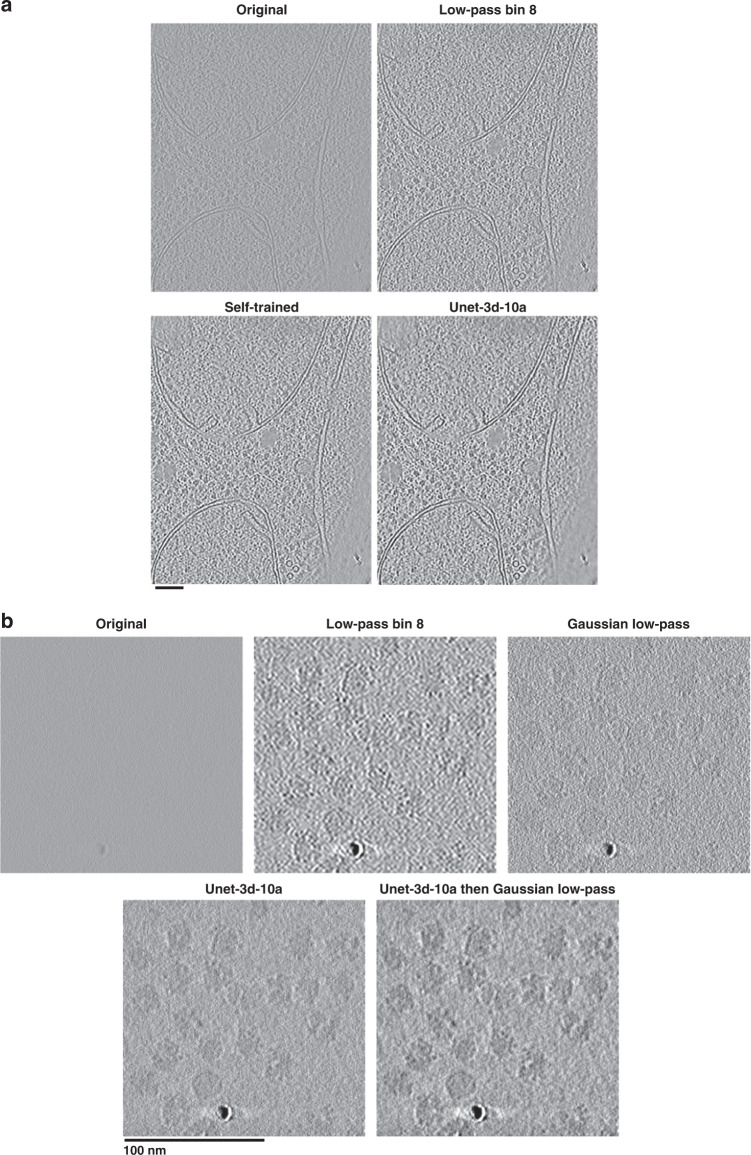
Fig. 4. Denoising with the 3D general model in Topaz improves cellular and single particle tomogram interpretability.
a Saccharomyces uvarum lamellae cryoET slice-through (collected at 6 Å pixelsize and 18 microns defocus, then binned by 2). The general denoising model (Unet-3d-10a) is comparable visually and by SNR to the model trained on the tomogram’s even/odd halves (Self-trained). Both denoising models show an improvement in protein and membrane contrast over binning by 8 while confidently retaining features of interest, such as proteins between membrane bilayers. Both denoising models also properly smooth areas with minimal protein/membrane density compared to the binning by 8. See Supplementary Movie 1 for the tomogram slice-throughs. b 80S ribosomes as single particles (EMPIAR-10045; collected at 2.17 Å pixelsize and 4 microns defocus). The general denoising model (Unet-3d-10a) is markedly improved over binning by 8 and the 1/8 Nyquist Gaussian low-pass, both with smoothing background appropriately while increasing contrast and with retaining features of interest at high fidelity, such as the RNA binding pocket in all orientations. The same 1/8 Nyquist Gaussian low-pass applied to the denoised tomogram further improves contrast by suppressing high-frequencies that the user may deem unimportant. See Supplementary Movie 2 for the tomogram slice-throughs.