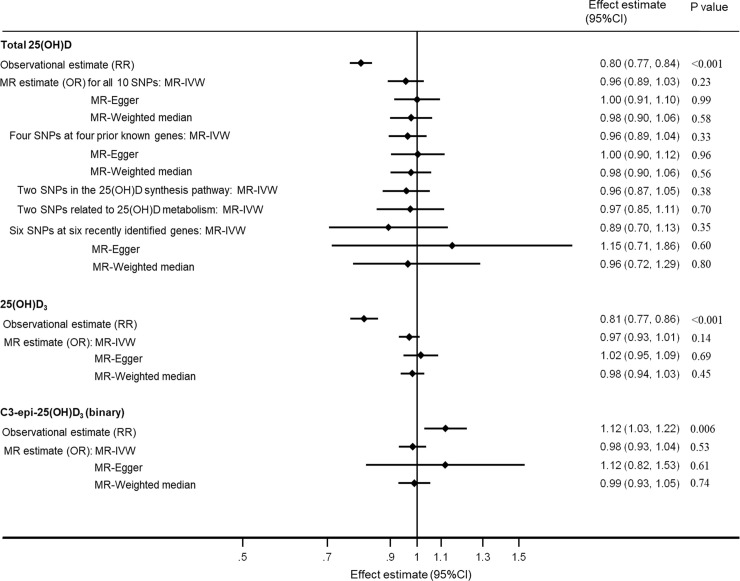
Fig 3. Association of 25(OH)D metabolites with type 2 diabetes from observational and MR analyses.
Estimates (95% CIs) were scaled to represent RRs from observational analyses or ORs from MR per 1–standard deviation difference in each 25(OH)D metabolite, except for the binary C3-epi-25(OH)D3 variable (above versus below the lower limit of quantification). For the MR sensitivity analysis for total 25(OH)D, the 4 prior known genes are GC, CYP2R1, NADSYN1/DHCR7, and CYP24A1; the 25(OH)D synthesis pathway genes are CYP2R1 and NADSYN1/DHCR7; the 25(OH)D metabolism genes are GC and CYP24A1; and the 6 recently identified genes are PADI1, CRCT1, UGT1A5, AMDHD1, SEC23A, and SULT2A1. For each analysis, 3 MR methods were used, including the IVW method, MR-Egger method, and weighted median method. For MR analyses based on 2 SNPs, the MR-Egger and weighted median methods could not be applied. For the observational estimate, the total number of type 2 diabetes cases and non-cases was 15,611 and 21,106, respectively, for total 25(OH)D, and 8,331 and 11,837, respectively, for 25(OH)D3 (or C3-epi-25(OH)D3). For the MR estimate of all 3 vitamin D variables, the total number of cases and non-cases was 80,983 and 842,909, respectively. 25(OH)D, 25-hydroxyvitamin D; CI, confidence interval; IVW, inverse-variance-weighted; MR, Mendelian randomisation; OR, odds ratio; RR, relative risk; SNP, single nucleotide polymorphism.