Abstract
Mushroom-forming fungi are complex multicellular organisms that form the basis of a large industry, yet, our understanding of the mechanisms of mushroom development and its responses to various stresses remains limited. The winter mushroom (Flammulina filiformis) is cultivated at a large commercial scale in East Asia and is a species with a preference for low temperatures. This study investigated fruiting body development in F. filiformis by comparing transcriptomes of 4 developmental stages, and compared the developmental genes to a 200-genome dataset to identify conserved genes involved in fruiting body development, and examined the response of heat sensitive and -resistant strains to heat stress. Our data revealed widely conserved genes involved in primordium development of F. filiformis, many of which originated before the emergence of the Agaricomycetes, indicating co-option for complex multicellularity during evolution. We also revealed several notable fruiting-specific genes, including the genes with conserved stipe-specific expression patterns and the others which related to sexual development, water absorption, basidium formation and sporulation, among others. Comparative analysis revealed that heat stress induced more genes in the heat resistant strain (M1) than in the heat sensitive one (XR). Of particular importance are the hsp70, hsp90 and fes1 genes, which may facilitate the adjustment to heat stress in the early stages of fruiting body development. These data highlighted novel genes involved in complex multicellular development in fungi and aid further studies on gene function and efforts to improve the productivity and heat tolerance in mushroom-forming fungi.
Introduction
Mushroom-forming fungi are widely distributed through Earth’s ecosystems. They play essential roles in nutrient cycling, environmental protection, plant and animal health [1–3]. Mushrooms are also important food sources and produce molecules with therapeutic activities and enzymes that can be applied in bioconversion [4, 5]. Furthermore, the fruiting body is a complex multicellular structure [6], whose complexity level resembles that of multicellular plants and animals. Therefore, understanding fruiting body development is important also from the perspective of understanding major evolutionary transitions.
Fruiting body development starts with the aggregation of aerial dikaryotic hyphae under suitable environmental conditions (nutrient, light, and temperature etc.) [7–13]. These aggregates continuously develop into primordia, which further differentiate into mature fruiting bodies [7–13]. Then, karyogamy and meiosis take place in the basidia within the hymenium of the fruiting body, and additional mitosis results in basidiospores [7–13]. Coprinopsis cinerea and Schizophyllum commune were used as the main model species to study the mechanisms of mushroom formation, due to their short life cycles and suitability for genetic manipulation [7–9]. Studies on these two model species using tools such as DNA-mediated transformation, RNA interference, and CRISPR/Cas9 etc. have pioneered our knowledge of the multicellular development, mating pheromone, and receptor signaling pathways in the Agaricomycetes [14–18]. More recently, studies also focused on ecologically or economically important non-model species, which included the saprotrophic fungi (Agaricus bisporus, Flammulina filiformis, Lentinula edodes, Lentinus tigrinus, Cyclocybe aegerita), plant pathogen (Armillaria ostoyae), and the ectomycorrhizal fungi (Laccaria bicolor) [2, 19–26]. These studies broadened our knowledge on fruiting body development and also highlighted conserved expression patterns of some key developmental genes (such as the genes encoding light receptors (white collar complex), transcription factors (c2h2, hom1, hom2), CAZyme and F-box protein etc.), indicating conserved molecular mechanisms in multicellular complexity in Agaricomycetes. However, mushroom development is a highly organized process, and genetic drivers of spatial and temporal differentiation events are not known, and our understanding of mushroom formation in other ecologically and economically important mushroom-forming fungi is still in its infancy [12, 13, 27, 28].
The winter mushroom or enokitake, Flammulina filiformis (formerly known as F. velutipes) [29], is cultivated at large scales in East Asia [30–32]. A comprehensive understanding of fruiting body development of this mushroom would not only benefit its production, but can also help to uncover conserved molecular mechanisms of development in the Agaricomycetes. Commercial scale cultivation of this mushroom requires a low temperature (≤15°C) (since the wild strain commonly fruiting during late autumn to early spring), which costs large amounts of energy, especially during summer in East Asia [29, 33, 34]. Fortunately, a heat resistant strain (called M1 after Mingjin1) has been isolated in subtropical areas (Fujian province) of China in summer that can fruit at 23°C and thus has great potentials in strain improvement and should be subject to studies of heat resistance [33].
Previous studies revealed molecular details in fruiting body development in Flammulina species at the transcriptome, proteome, single gene or protein level [20, 35–39]. A series of genes associated with mushroom formation, including the mating type genes, hydrophobins, and fruiting body specific genes have been identified [20, 31, 40]. Researches also investigated at least two genes controlling fruiting at >15°C based on hybridization analysis [34]. However, the molecular response to heat stress, in particular those of the well-known heat shock protein coding gene hsp70, hsp90 and other molecular chaperons etc., in F. filiformis remains unknown. Although previous studies provided us with a basic understanding of fruiting body development of F. filiformis or its closely related F. velutipes, our knowledge about the fruiting body development and heat stress response of this mushroom is still incomplete.
In this study, we aimed to (i) uncover key fruiting body genes in various developmental stages; (ii) investigate whether conserved developmental patterns exist in F. filiformis and other Agaricomycetes; and (iii) understand the responses to heat stress of heat sensitive (XR) and resistant (M1) strains and identify the key heat stress response genes in F. filiformis. We sampled RNA in different developmental stages of the M1 strain, and from the primordium stage grown at 10°C and 18°C of both the M1 and XR strains, for studying fruiting body development and its response to heat stress, respectively. Our results identified conserved gene expression patterns of fruiting body development in the Agaricomycetes and revealed that the heat tolerant strain M1 differentially expressed more genes in response to heat stress than the heat sensitive strain XR.
Materials and methods
Strains and culture conditions
The heat tolerant strain M1 (CGMCC5.2219) was domesticated from a wild strain collected in subtropical areas in China in summer (Fujian province). The heat sensitive strain XR (CGMCC5.2218) was isolated from a mushroom market, this strain was imported from Japan [33]. Both of them are deposit in the Chinese General Microbiological Culture Collection Center (CGMCC). They were grown on enriched Potato Dextrose Agar (PDA) medium (0.05% KH2PO4, 0.05% MgSO4, 2% glucose, 0.2% yeast extract, 0.2% peptone, 1.8% agar) in 90 mm Petri dish for 10 days at 23 °C. Then, the mycelium was inoculated in liquid cultures in 500ml Erlenmeyer flask containing 200ml enriched Potato Dextrose Broth (PDB) medium (0.05% KH2PO4, 0.05% MgSO4, 2% glucose, 0.2% yeast extract, 0.2% peptone), shaken at 150 r.p.m for 10 days at 23 °C. Afterwards, the mycelium was inoculated to a growth medium consisting of 90% cottonseed hull, 10% wheat bran, and 65% water in 1100 ml disposable bottles after sterilization. Inoculated bottles were incubated at 23 °C under dark conditions for 30 days, and then the mycelium scratched to emulates disturbance and transferred to 23 °C with 95% humidity for fruiting.
Sample collection for RNA-seq
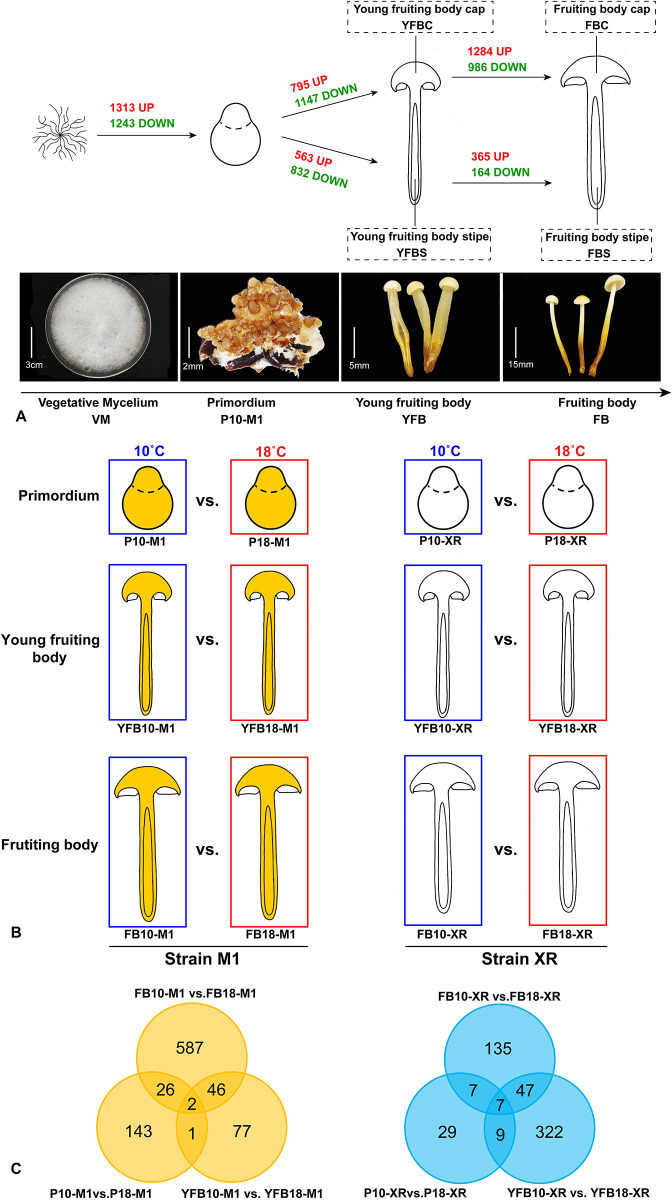
We selected different developmental stages of strain M1 grown at 10 °C (except for the mycelium, which was grown at 23 °C). We collected the vegetative mycelium (VM), the primordium (P10-M1), the young fruiting body cap (YFBC), the fruiting body cap (FBC), the young fruiting body stipe (YFBS), and the fruiting body stipe (FBS) (Fig 1A). For the heat stress response study, we collected the primordia, young fruiting body, and fruiting body of M1 strain grown at 18 °C (P18-M1), as well as the primordia, young fruiting body and fruiting body of XR strain grown at 10 °C and 18 °C respectively (Fig 1B).
Fig 1. Overview of the sampling and differential expression analyses used in this study.
A: fruiting body development. B: heat stress; C: Venn diagram of the numbers of up-regulated genes at 18°C compared to 10°C in each developmental stage of the M1 and XR strain.
Total RNA preparation and transcriptome sequencing
Samples from three biological replicates were flash-frozen in liquid nitrogen and stored at -80 °C. Total RNA of each sample was extracted using the RNAprep Pure toolkit, following the manufacturer’s protocol (TIANGEN, Beijing, China). Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform and 125 bp/150 bp paired-end reads were generated. All raw-sequence reads data were deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with accession number of PRJNA557510.
Read mapping to the reference genome, FPKM and gene annotation
Clean data were obtained by removing reads containing adapters, reads containing poly-N and low-quality reads from raw data through Trimmomatic (v.0.33) [41]. Then, we used HISAT2 (v.2.10) [42] to map the clean reads to the reference genome which assembled to chromosome level [20], and employed StringTie (v.1.3.4) [43] to calculate each gene’s FPKM value. All of the genes were annotated using local BLASTX programs against the Nr, SwissPort, GO and PFAM databases.
CAZyme gene annotation
Carbohydrate-active enzymes (CAZymes) were classified separately by HMM search of dbCAN HMMs 4.0 [44] (default cutoff threshold) and BLASTP search of the CAZy database [45] (evalue < = 1x10-6 and coverage > = 0.2, maximum hit number is 500).
Differential gene expression analysis
We performed differential gene expression analyses on each adjacent developmental stage of the M1 strain (Fig 1A) as well as the primordium stage grown at 10 °C vs 18 °C of the M1 and XR strains (Fig 1B). Analyses were based on three biological replicates per condition and were performed using the DESeq package (1.18.0) [46]. The input data of young fruiting body grown at 10 °C of M1 (YFB10-M1) were used as the pooled reads of young fruiting body cap (YFBC) and young fruiting body stipe (YFBS) of M1, and the input data of fruiting body grown at 10°C of M1 (FB10-M1) were used as the pooled reads of fruiting body cap (FBC) and fruiting stipe (FBS) of M1. Genes with log2 (fold change) ≥ 1 and Padj ≤ 0.05 were considered as differentially expressed gene (DEG).
Gene network construction and visualization
Co-expression networks were constructed using the WGCNA package in R [47]. Genes with averaged FPKM from three replicates higher than 1 in at least one sample were input to WGCNA unsigned co-expression network analysis (S1 Table). The modules were obtained using the step-by-step network construction function on block-wise modules with default settings, except that the power is 24 for fruiting body development analysis, 20 for M1 and 10 for XR in heat stress response analysis. TOM-Type was set to signed, minModuleSize to 30, and mergeCutHeight to 0.25. The networks were visualized using Cytoscape (v.3.5.1) [48].
Comparative genomic analysis
In order to check the conservation level of the developmentally regulated genes? during fruiting body development, analyzed a 201 genome dataset (ranging from unicellular yeasts to filamentous and complex multicellular fungi which also included F. filiformis in this study) and the corresponding phylogenetic tree taken from a previous study [13]. Conservation of genes was assessed based on the phylogeny, by assessing the presence/absence of genes across the panel of species.
qRT-PCR analysis
Reverse transcription of RNA (1ug) in a 20 μL reaction volume was performed using TUREscript 1st Stand cDNA SYNTHESIS Kit, following the manufacturer’s protocol (Aidlab, Beijing, China). Reactions were incubated at 42 °C for 60min, and 65 °C for 10min. The amplifications were performed using 5 μL SYBR qPCR Mix, 0.5 μL forward primer, 0.5 μL reverse primer and 1 μL cDNA, and 3 μL ddH2O in a final volume of 10 μL. The cycling parameters were 95 °C for 3 min followed by 30 cycles of 95 °C for 10 s, 58 °C for 30 s and 72 °C for 20 s. The relative gene expression was analyzed calculated by the qPCRsoft3.2. The 18S ribosomal RNA gene was used as the internal reference. The primers of each gene were listed in S2 Table.
Results and discussion
Overview of the transcriptome sequence data
We obtained 39.4–63.6 million paired-end reads for 15 sample types in triplicates (45 libraries in total) (S3 Table). Of the quality-filtered reads 58.3–71.2% mapped to the reference genome of F. filiformis (S3 Table). The moderate mapability may be caused by the different strains used in this study compared with the reference genome from strain KACC42780 [20]. Although we expect this to not influence our results, it may cause lower sensitivity in faster evolving or strain specific genes. Differences in read mapability were not found between the M1 and XR strain which made the transcriptome comparable across these two strains (S3 Table). To validate the results of the RNA-seq analysis, 18 genes were randomly selected for quantitative real-time PCR (qRT-PCR). These genes showed expression patterns similar to those in the RNA-seq data (S1 Fig), indicating that our transcriptome sequencing provided a good estimate of gene expression patterns in the analyses of fruiting body development and heat stress response of F. filiformis.
Temporal- and spatial-gene expression across F. filiformis development
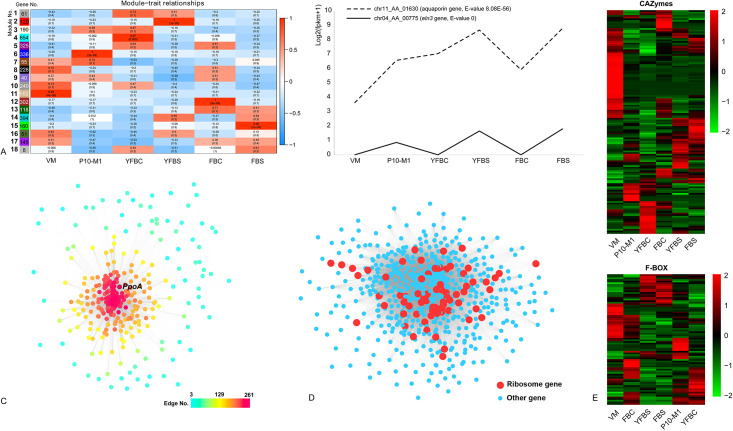
Differential expression analysis indicated the highest number of differentially expressed genes (DEGs) in the transition from vegetative mycelium to primordium (1,313 up-regulated, 1,243 down-regulated), followed by young to mature fruiting body cap (1,284 up-regulated, 986 down-regulated) relative to primordium (Fig 1A; S2A Fig, S4 Table). This gene expression pattern was also recognized by the WGCNA analysis, a systems biology approach aimed at uncover gene modules which share gene expression patterns at a pre-specified similarity cutoff [47]. We identified six gene modules highly correlated with a single tissue type (Fig 2A; S5 Table). Among them, the primordium module (module no. 6), young fruiting body cap module (module no. 4) and mature fruiting body cap module (module no. 12) contained the highest number of genes (Fig 2A). These results indicate that primordium stage comprises the most significant morphogenetic transition, and that hymenium maturation and sporulation in young and mature fruiting body caps may also harbor complex molecular mechanisms. This gene expression pattern is consistent with those found in other Agaricomycetes [8, 22, 13, 49]. Moreover, the DEGs related to each developmental stage were enriched for GO terms typical for fruiting body formation, see S3 Fig and S6 Table.
Fig 2. Gene expression patterns in fruiting body development of F. filiformis.
A: Gene module-sample association revealed by gene co-expression analysis in WGCNA. Each row corresponds to a module, each column corresponds to a developmental stage or tissue type. VM, P10-M1, YFBC, YFBS, FBC and FBS correspond to vegetative mycelium, primordium, young fruiting body cap, young fruiting body stipe, fruiting body cap and fruiting body stipe, respectively. Upper and lower numbers in the cells indicate the correlation coefficient between the module and sample and the significance of the correlation (p-value), respectively; B: Expression patterns of the three aquaporin genes and the eln3 gene in each developmental stage of strain M1. E-value was reported by BLASTX search; C: Gene co-expression networks of the primordium module (module no. 6). The scale bar indicates the number of connections a gene has; D: Gene co-expression network of young fruiting body cap module (module no. 4). The red dots represent ribosomal protein encoding gene, blue dots represent other gene; E: Heatmap of the CAZyme and F-BOX gene expression in each developmental stage.
Primordium development includes genes widely conserved in Agaricomycetes
The top 20 up-regulated genes induced in primordium relative to vegetative mycelium were listed in Table 1. Building on a previously published dataset [13], we found these genes were widely conserved in fungi, and re-emphasized that several primordium-upregulated genes have homologs in simple multicellular or unicellular fungi. This indicates that some conserved gene families were recruited for complex multicellularity during evolution [13]. Among them, the Gti1/Pac2 family is conserved in all fungi (Table 1; Fig 3). Previously, this family has been discussed mostly in yeasts and pathogenic fungi, where it plays an important role in fungal growth and development [50]. Recent research revealed that this family is also developmentally regulated in Armillaria ostoyae, Coprinopsis cinerea, Lentinus tigrinus, Rickenella mellea, Schizophyllum commune, and Phanerochaete chrysosporium [13]. Thus, these genes may also play a key role in fruiting body development in Agaricomycetes. In addition, three TFs in the top 20 up-regulated genes (Zinc finger, C2H2 type, Zinc finger, Ring type, and Zn (2)-C6 fungal type) are conserved in Dikarya and Zoopagomycota plus later diverging phyla (Table 1; Fig 3), which reinforces the role of these TFs in complex multicellularity in fungi [13].
Table 1. Top 20 most upregulated and two other notable genes up-regulated in the primordium stage relative to vegetative mycelium.
Aquaporin and hydrophobin genes mentioned in the text are also shown.
| Rank | Protein ID | Log2 (FC) | P-value (FC) | Best Hit (Accession No.) | |
|---|---|---|---|---|---|
| 1 | chr11_AA_00208 | 10.37 | 5.8x10-14 | - | |
| 2 | chr08_AA_01205 | 9.39 | 5x10-55 | Hypothetical protein | |
| 3 | chr10_AA_00968 | 8.87 | 4.3x10-60 | Short-chain dehydrogenase (IPR002347) | |
| 4 | chr11_AA_00046 | 8.70 | 1.9x10-138 | Gti1/Pac2 family (IPR018608) | |
| 5 | chr10_AA_00489 | 8.65 | 5x10-12 | Flammutoxin (BAA32792) | |
| 6 | chr01_AA_00267 | 8.62 | 7.4x10-25 | - | |
| 7 | chr03_AA_00235 | 8.52 | 7.5x10-30 | - | |
| 8 | chr08_AA_01207 | 8.37 | 7.9x10-16 | - | |
| 9 | chr08_AA_01206 | 8.35 | 1.3x10-13 | - | |
| 10 | chr11_AA_00874 | 8.28 | 5.2x10-18 | Schizophyllum commune hydrophobin, Sc3 (P16933) | |
| 11 | chr11_AA_01512 | 8.24 | 9.2x10-7 | Cytochrome P450 (IPR001128) | |
| 12 | chr07_AA_00932 | 8.22 | 6x10-21 | Zinc finger, RING-type (IPR001841) | |
| 13 | chr03_AA_00276 | 7.97 | 8.1x10-95 | Flammulina velutipes hydrophobin, fv-hyd1 (AB126686) | |
| 14 | chr04_AA_00509 | 7.91 | 5.4x10-85 | Zinc finger, C2H2 (IPR007087) | |
| 15 | chr05_AA_00568 | 7.77 | 6.2x10-126 | Kre9/Knh1 family (IPR018466) | |
| 16 | chr08_AA_00570 | 7.01 | 1x10-127 | - | |
| 17 | chr01_AA_00480 | 7.68 | 1.4x10-17 | - | |
| 18 | chr08_AA_01181 | 7.56 | 1.3x10-117 | - | |
| 19 | chr09_AA_01255 | 7.53 | 1.2x10-26 | Zn(2)-C6 fungal-type (IPR001138) | |
| 20 | chr10_AA_01153 | 7.47 | 9x10-17 | - | |
| 47 | chr11_AA_01264 | 5.81 | 3x10-79 | Aquaporin (P43549) | |
| 54 | chr05_AA_00590 | 5.63 | 1x10-9 | Flammulina velutipes hydrophobin, fv-hyd1 (AOV80987) |
FC: Fold Change; Chytridio: Chytridiomycota; Mucoro: Mucoromycota; Zoopago: Zoopagomycota.
Genes conservation level in Fungi
Black-Species specific
Blue-Conserved in Physalacriaceae
Light blue-Conserved in Agaricomycetes
Green-Conserved in Basidiomycota
Yellow-Conserved in Dikarya
Orange-Conserved in Chytridio/Mucoro/Zoopago+higher
Red-Conserved in Fungi
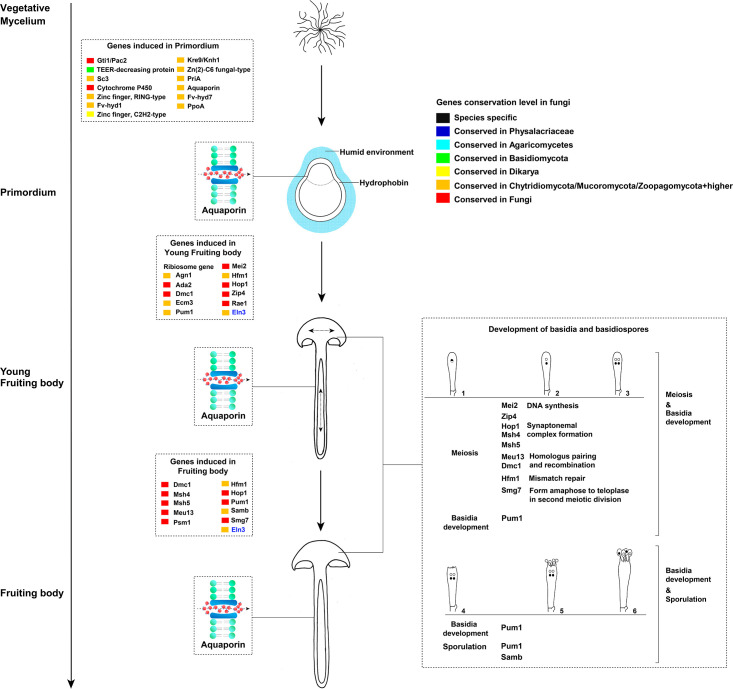
Fig 3. Synoptic summary of key genes at different stages of development of F. filiformis.
1–6 indicates developmental stages of basidia.
We found the gene encoding flammutoxin was conserved in 19 species in Agaricomycetes and 1 species in the Dacrymycetes (Table 1; Fig 3). This protein has been studied in F. filiformis, and may form a pore in the intestinal epithelial cells of fungivorous animals, leading to cell death [51]. However, because this protein is heat-labile, clinical reports about the intestinal dysfunction caused by ingestion of this mushroom are rare. Pore-forming proteins have been studied in Pleurotus species, which exhibit cytotoxicity toward insect cells via pore formation in cell membranes to defend predation [52, 53]. Thus, whether flammutoxin serves to protect the fruiting body of F. filiformis predated by mammals or insects needs further investigation. In addition, a ricin-B lectin gene (chr11_AA_01461), showed high expression in fruiting bodies. It is homologous to the Macrolepiota procera mpl, Mpl protein shows toxicity towards the nematode Caenorhabditis elegans [54]. A re-analysis of its homologs in C. cinerea, L. tigrinus, R. mellea, S. commune, and P. chrysosporium revealed that it has fruiting body specific expression pattern in these species as well. This is in concordance with its expression pattern in M. procera, indicating a conserved role of this gene in protecting fruiting bodies against predators and parasites in Agaricomycetes [54]. Compared with the conserved flammutoxin and ricin-B lectin gene discussed here, we found the previously mentioned widely conserved ribotoxins gene is not exists in F. filiformis genome [55].
Interestingly, the second most up-regulated gene in primordium was restricted to the family Physalacriaceae, which indicated a role in primordium formation in this family (Table 1; Fig 3). Unfortunately, our knowledge about this gene is limited. However, recent study reported the successful overexpression and RNA interference of the transcription factor pdd1 in F. filiformis, which provides genetic tools to study this gene in the future [56]. Based on our analysis, almost half of the top 20 up-regulated genes in primordium stage without annotations (Table 1), they are widely conserved in simple multicellular and complex multicellular fungi, indicating that systematic studies are needed on mushroom development to help to understand multicellularity.
Notable genes during fruiting body development
In the primordium stage, we found that genes encoding hydrophobins were (homologs of fv-hyd1, fv-hyd7 and S. commune sc3) significantly up-regulated relative to vegetative mycelium (Fig 3; S2A Fig; S4 Table). It is well known that hydrophobins and cerato-platanins assemble at the hyphae surface to promote their aggregation in humid environments [57–59]. Hydrophobins may also hinder water absorption through the membrane [60].
We detected the aquaporin gene specifically induced in primordia and stipe tissues (Fig 2B; S2A Fig; S4 Table). Aquaporins are integral membrane proteins responsible for water and solute transport, and also involved in mycorrhizal formation and plant-fungal interactions during symbiosis establishment [26, 61]. Recent study revealed that aquaporins were also developmentally regulated in L. bicolor fruiting bodies [26]. However, aquaporins have been discussed mostly in the context of mycorrhizal fungi [62–65]. Based on our results, it seems likely that aquaporin-dependent water transport is a key process during mushroom development in saprotrophic fungi too, possibly in water transport along the stipe to facilitate water supply of the developing cap and gills [64]. A re-analysis of the data published by Sipos et al. (2017) [22] and Krizsán et al. (2019) [13] indicated that aquaporins were developmentally regulated in all six species studied by these authors, indicating that the role of aquaporins in development is not restricted to Flammulina, but may be widely conserved in fruiting body development. Aquaporins were differentially expressed in mycorrhizal species [2, 26, 64], which provides additional support to the hypothesis that fruiting bodies and ectomycorrhizae have many shared gene expression patterns, possibly pointing to common developmental origins [2, 64, 66].
Interestingly, we found that the Flammulina homolog of the stipe elongation gene eln3 of C. cinerea possessed similar expression pattern to those of aquaporin genes during fruiting body development (Figs 2B and 3; S2A Fig; S4 Table). The mutant strain of this gene in C. cinerea produced aberrant fruiting bodies, in which the stipe hardly elongated during development [67]. The eln3 homolog of Volvariella volvacea was also reported to be differentially expressed during fruiting body development [68]. A re-analysis indicated that eln3 in C. cinerea and its homologs in L. tigrinus, A. ostoyae and R. mellea were developmentally regulated in RNA-Seq on data from previous studies [22, 13]. Homologs in S. commune and P. chrysosporium were not developmentally expressed, which might be explained by the lack of a stipe in these species. The broad conservation and expression patterns of eln3 suggests that the molecular mechanisms of stipe elongation may be shared in Agaricomycetes, despite the independent origins of pileate-stipitate fruiting bodies in the class [28]. These results further highlight this gene as an interesting target in future strain improvement programs.
We found that one of the hub genes in the primordium module (module no. 6, WGCNA co-expression analysis) was a homolog of Aspergillus nidulans ppoA (Figs 2C and 3). This gene participates in oxylipin synthesis, which modulates sexual and asexual development in A. nidulans [69]. During sexual development, the PpoA protein initially localized in Hülle cells formed at the stage of cleistothecial primordium formation, and subsequently in immature cleistothecia in A. nidulans [69]. Over-expression of this gene in A. nidulans promotes sexual spore formation [69]. A re-analysis of this gene’s homologs in C. cinerea, R. mellea, and A. ostoyae revealed a conserved expression pattern in these species, which indicates that oxylipins may mediate sexual development in the Ascomycota and the Agaricomycetes.
The homolog of Lentinula edodes priA was highly induced in primordium and young fruiting body cap of F. filiformis (S2A Fig; S4 Table). This gene was reported to possess the higher expression level in primordium and young fruiting body of L. edodes and over-expressing the priA gene in L. edodes monokaryotic mycelium remarkably decreased zinc ion accumulation, which indicates this gene may play a role in regulation of the intracellular zinc concentration [70]. Surprisingly, we found its homologs were highly expressed in vegetative mycelium and lower expressed in fruiting bodies in A. ostoyae, C. cinerea, L. tigrinus, R. mellea, S. commune, and P. chrysosporium.
A large number (74) of ribosomal protein encoding genes, and homologs of genes involved in cell differentiation and cell wall formation in S. pombe (agn1 and rae1), Candida albicans (ada2), and S. cerevisiae (ecm3) were hub genes in the young fruiting body cap module of the WGCNA analysis or were up-regulated relative to the primordium stage (Figs 2D and 3; S2A and S2B Fig; S4 Table). These results might reflect intense growth and protein synthesis in young fruiting bodies. Although the hymenium was immature in this stage, four meiosis regulation genes homologous to S. pombe mei2, Cryptococcus neoformans dmc1, and S. cerevisiae hfm1, hop1, and zip4 were up-regulated (Fig 3; S2A Fig; S4 Table). Among them, dmc1, hop1 and hfm1 homologs were also up-regulated in mature cap (Fig 3; S2A Fig; S4 Table).
Compared to young fruiting body cap, more meiosis genes (homologs of S. pombe psm1 gene, C. neoformans dmc1 gene, and S. cerevisiae, msh4, msh5, meu13, hop1, hfm1, and smg7) were induced in the fruiting body cap (Fig 3; S2A Fig; S4 Table). Among the genes induced in young fruiting body cap and fruiting body cap, the genes homologous to C. neoformans pum1 may be noteworthy (Fig 3; S2A Fig; S4 Table). Pum1 is an RNA binding protein, and possesses an important role in post-transcriptional regulation in basidium development and sporulation in C. neoformans [71–73]. Previous studies revealed that the knockout of this gene in C. neoformans resulted in a severe defect in basidium formation [71–73]. In this study, we detected five genes homologous to C. neoformans pum1, two of them were developmentally regulated in young fruiting body cap and fruiting body cap, which indicates they may participate in basidium formation and sporulation in F. filiformis (Fig 3; S2A Fig). We detected another sporulation-related gene, homologous to A. nidulans samB (Fig 3; S2A Fig). Knock out of this gene in A. nidulans hindered ascospore formation [74]. A re-analysis of the homologs of pum1 and Samb in A. ostoyae, C. cinerea, L. tigrinus, R. mellea, S. commune, and P. chrysosporium revealed they possess conserved expression patterns in these species. Due to these two genes were widely conserved (Fig 3), we therefore speculate that some molecular mechanisms of spore formation may be conserved in fungi.
CAZymes and F-box genes
Certain CAZymes were shown or assumed to participate in cell wall remodeling during fungal tissue differentiation [13, 49, 75–77]. We annotated 407 CAZymes genes in F. filiformis, 137 of them were differentially expressed (Fig 2E; S7 Table), which is consistent with previous studies in other mushroom-forming fungi [13, 20, 49]. Among these genes, Glycoside hydrolases (GH) and Glycosyltransferases (GT) were most abundant, with 57 and 30 genes, respectively. Although the targets of these families in fruiting bodies are currently unknown, the stage specific expression of these genes during fruiting body development reinforces the view that cell wall remodeling is a widespread and well-organized process in fruiting body development in Agaricomycetes.
F-box proteins play a key role in protein ubiquitination and modification, and are involved in many important biological processes not only in plants, but all eukaryotic [78, 79]. They were recently reported in relation to fruiting body development [13, 49]. In this study, 210 F-box encoding genes were annotated in F. filiformis, of which 80 were developmentally expressed and showed stage-specific expression patterns (Fig 2E; S8 Table). Similar expression patterns were also recognized in A. ostoyae, C. cinerea, R. mellea, L. tigrinus, and S. commune, which suggests that F-box genes may be crucial during fruiting body development in Agaricomycetes [13].
Strain M1 expressed a large gene pool in response to heat stress relative to XR
Based on cultivation tests, we found that the growth of M1 strain showed no difference in 10°C and 18°C, while, the growth in XR strain was obviously retarded at 18 °C. On the molecular level, we found more differentially expressed genes in M1 (882 DEGs) than in XR (556 DEGs) (Fig 1C; S4 Fig). Based on the Venn diagram on Fig 1, we found two genes with elevated expression level in all developmental stages of strain M1 at 18 °C: a DNA damage repair gene homologous to S. cerevisiae rad18 and a epoxide hydrolase gene homologous to A. niger. For strain XR, seven genes had an elevated expression level at all developmental stages under 18 °C (Fig 1C): two hsp20 genes, one WD repeat-containing gene, and four genes without annotation. Consistent with these functions, a GO enrichment analysis revealed that up-regulated genes in M1 were mainly enriched in ‘response to stress’ (GO:0006950, P<0.05), ‘protein folding’ (GO:0006457, P<0.01), and ‘chaperone binding’ (GO:0051087, P<0.05) etc. And the genes up-regulated in XR were mainly enriched in ‘protein binding’ (GO:0005515, P<0.01), ‘DNA binding’ (GO:0003677, P<0.01), and ‘protein kinase binding’ (GO:0019901, P<0.01) etc. (S5 Fig; S9 Table). These results indicate the different heat stress response strategies were employed in these two strains.
Specifically, differential expression analyses revealed that the M1 strain had more heat shock protein genes up-regulated than XR (S4 Fig). Among them, homologs of S. cerevisiae hsp70, S. pombe hsp90 and another gene homologous to Ustilago maydis fes1 induced in M1 at 18°C may be noteworthy. The Hsp70 protein could protect nascent polypeptides and refold the damaged proteins under heat stress conditions [80]. If protein folding fails with Hsp70, Fes1 could interact with misfolded proteins and lead to their destruction by the ubiquitin-proteasome machinery [81]. Compared with Hsp70, Hsp90 functions primarily in the final maturation of proteins. Therefore, these genes may act as an “assembly line” [80] of protein maturation under heat stress during primordium development of strain M1. The heat stress induction of hsp70 and hsp90 was also reported in Lentinula edodes and Ganoderma lucidum [82, 83]. Homologs of these two genes were not differentially expressed in XR strain. Instead, the genes homologous to S. pombe hsp20 were up-regulated in XR strain in all developmental stages at 18°C, they play different roles than hsp70 and hsp90, which are probably required to prevent misfolded protein aggregation and their degradation under heat stress [80].
In addition, the DNA damage repair gene homologous to S. cerevisiae rad18 was up-regulated in all developmental stages in M1. Yeast strains lacking Rad18 proteins may be highly sensitive to a wide variety of DNA damaging agents such as UVC-light, ROS stress and γ-radiation [84–86]. However, this gene was only up-regulated in the young fruiting body stage of XR grown at 18°C. This example, combined with the fewer DEGs in the XR strain may indicate the loss of an ancestral heat stress response mechanisms in the commercial XR strain. This may result from the lack of temperature fluctuations and stress in general in factory settings.
Conclusions
This study broadened our knowledge of fruiting body development and heat stress response of mushroom-forming fungi based on comparisons of transcriptomic data in F. filiformis. We detected a series of genes (e.g. aquaporins, eln3-homologs, hydrophobins, conserved transcription factors, oxylipin biosynthesis genes) that show conserved, dynamic expression during fruiting body development, and also uncovered signal for defense against high temperature in the heat tolerance strain (M1) (e.g. hsp70, hsp90 and fes1 homologs). These, or other differentially expressed genes, might be good candidates for in-depth experimental follow-up analyses (e.g. gene knockout) to understand their specific roles and answer important or interesting questions that remained open. Analyzing the function of conserved genes in model and non-model species will be necessary to broaden our knowledge on fruiting body development in the Agaricomycetes.
Supporting information
VM, P10-M1, P18-M1, YFBC, YFBS, FBC and FBS correspond to vegetative mycelium, primordium grown at 10°C, primordium grown at 18°C, young fruiting body cap, young fruiting body stipe, fruiting body cap and fruiting body stipe, of strain M1. P10-XR, P18-XR correspond to primordium grown at 10°C and 18°C of strain XR. Bar chart represents the FPKM values (left vertical axis), line chart represents the real-time PCR expression values (right vertical axis).
(JPG)
A: Volcano plots of differential expression analysis for each comparison group; B: Gene co-expression network of the young fruiting body cap module (module no. 4 in Fig 2A). The scale bar indicates the number of connections a gene has.
(JPG)
X-axis indicates the ratio of the number of test genes and reference genes; Y-axis indicates the description of the functional terms.
(JPG)
(JPG)
X-axis indicates the ratio of the number of test genes and reference genes; Y-axis indicates the description of the functional terms.
(JPG)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
The authors are grateful to Prof. Won-Sik Kong (Mushroom Research Division, National Institute of Horticultural and Herbal Science, Rural Development Administration, Eumsung, Republic of Korea) for generously sharing the genome sequencing data of the strain KACC42780 for us. Ms. Ming-Li Li (Kunming Institute of Zoology, Chinese Academy of Sciences), Dr. Botond Hegedüs and Dr. Balázs Bálint (Synthetic and Systems Biology Unit, Institute of Biochemistry, Biological Research Centre, Szeged), Dr. Ti-Cao Zhang (Yunnan University of Chinese Medicine), Dr. Peter Langfelder (University of California), Dr. Yong-Ping Fu (Jilin Agricultural University) and Dr. Robin Ohm (Utrecht University) were acknowledged for their kind help in data analyses. The anonymous reviewers are also gratefully acknowledged for their comments and suggestions.
Data Availability
All raw reads were deposited in NCBI Sequence Read Archive (SRA,http://www.ncbi.nlm.nih.gov/Traces/sra) with accession number of PRJNA557510.
Funding Statement
This study was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (No. XDB31000000); Yunnan Ten-Thousand-Talents Plan - Yunling Scholar Project; and the National Basic Research Program of China (973 Program, No. 2014CB138305). LGN acknowledges support from the European Research Council (grant no. 758161 to L.G.N.) and the National Research, Development and Innovation office (Contract No. Ginop-2.3.2-15-00001, to LGN).
References
- 1.Xu J. Fungal DNA barcoding. Genome. 2016;59: 913–932. 10.1139/gen-2016-0046 [DOI] [PubMed] [Google Scholar]
- 2.Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452: 88 10.1038/nature06556 [DOI] [PubMed] [Google Scholar]
- 3.Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagye LG, et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. U. S. A. 2012;109: 17501–17506. 10.1073/pnas.1206847109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lomascolo A, Stentelaire C, Asther M, Lesage-Meessen L. Basidiomycetes as new biotechnological tools to generate natural aromatic flavours for the food industry. Trends Biotechnol. 1999;17: 282–289. 10.1016/s0167-7799(99)01313-x [DOI] [PubMed] [Google Scholar]
- 5.Ohm RA, de Jong JF, de Bekker C, Wösten HAB, Lugones LG. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011;81: 1433–1445. 10.1111/j.1365-2958.2011.07776.x [DOI] [PubMed] [Google Scholar]
- 6.Nagy LG, Gábor MK, Krisztina K. Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biol. Rev. 2018;93: 1778–1794. 10.1111/brv.12418 [DOI] [PubMed] [Google Scholar]
- 7.Kües U. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 2000;64: 316–353. 10.1128/mmbr.64.2.316-353.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohm RA, De Jong JF, Lugones LG, Aerts A, Kothe E, Stajich JE, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010;28: 957 10.1038/nbt.1643 [DOI] [PubMed] [Google Scholar]
- 9.Stajich JE, Wilke SK, Ahrén D, Au CH, Birren BW, Borodovsky M, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. U. S. A. 2010;107: 11889–11894. 10.1073/pnas.1003391107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kües U, Navarro-Gonzalez M. How do Agaricomycetes shape their fruiting bodies? 1. Morphological aspects of development. Fungal Biol. Rev. 2015;29: 63–97. [Google Scholar]
- 11.Muraguchi H, Umezawa K, Niikura M, Yoshida M, Kozaki T, et al. Strand-specific RNA-seq analyses of fruiting body development in Coprinopsis cinerea. PLoS One. 2015;10: e0141586 10.1371/journal.pone.0141586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto Y. Influences of environmental factors on fruiting body induction, development and maturation in mushroom-forming fungi. Fungal Biol. Rev. 2018;32: 236–248. [Google Scholar]
- 13.Krizsán K, Almási E, Merenyi Z, Sahu N, Viragh M, Koszo T, et al. Transcriptomic atlas of mushroom development reveals conserved genes behind complex multicellularity in fungi. Proc. Natl. Acad. Sci. U. S. A. 2019;116: 7409–7418. 10.1073/pnas.1817822116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binninger DM, Skrzynia C, Pukkila PJ, Casselton L. DNA‐mediated transformation of the basidiomycete Coprinus cinereus. The EMBO Journal. 1987;6: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wälti MA, Villalba C, Buser RM, Grünler A, Aebi M, Künzler M. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryot. Cell. 2006;5: 732–744. 10.1128/EC.5.4.732-744.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vonk PJ, Escobar N, Wösten HA, Lugones LG, Ohm RA. High-throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre-assembled Cas9 ribonucleoproteins. Sci. Rep. 2019;9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugano SS, Suzuki H, Shimokita E, Chiba H, Noji S, Osakabe Y, et al. Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci. Rep. 2017;7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelkmans JF, Patil MB, Gehrmann T, Reinders MJ, Wösten HA, Lugones LG. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci. Rep. 2017;7: 310 10.1038/s41598-017-00483-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehrmann T, Pelkmans JF, Ohm RA, Vos AM, Sonnenberg AS, Baars JJ, et al. Nucleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regulatory programs. Proc. Natl. Acad. Sci. U. S. A. 2018;115: 4429–4434. 10.1073/pnas.1721381115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park YJ, Baek JH, Lee S, Kim C, Rhee H, Kim H, et al. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PloS One. 2014;9: e93560 10.1371/journal.pone.0093560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelkmans JF, Vos AM, Scholtmeijer K, Hendrix E, Baars JJ, Gehrmann T, et al. The transcriptional regulator c2h2 accelerates mushroom formation in Agaricus bisporus. Appl. Microbiol. Biotechnol. 2016;100: 7151–7159. 10.1007/s00253-016-7574-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sipos G, Prasanna AN, Walter MC, O’Connor E, Bálint B, Krizsán K, et al. 2017. Genome expansion and lineage-specific genetic innovations in the forest pathogenic fungi Armillaria. Nat. Ecol. Evol. 2017;1: 1931 10.1038/s41559-017-0347-8 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Zeng X, Liu W. De novo transcriptomic analysis during Lentinula edodes fruiting body growth. Gene. 2018;641: 326–334. 10.1016/j.gene.2017.10.061 [DOI] [PubMed] [Google Scholar]
- 24.Wu B, Xu Z, Knudson A, Carlson A, Chen N, Kovaka S, et al. Genomics and development of Lentinus tigrinus: A white-rot wood-decaying mushroom with dimorphic fruiting bodies. Genome Biol. Evol. 2018;10: 3250–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herzog R, Solovyeva I, Bölker M, Lugones LG, Hennicke F. Exploring molecular tools for transformation and gene expression in the cultivated edible mushroom Agrocybe aegerita. Mol Genet Genomics. 2019;294: 663–677. 10.1007/s00438-018-01528-6 [DOI] [PubMed] [Google Scholar]
- 26.Xu H, Navarro-Ródenas A, Cooke JE, Zwiazek JJ. Transcript profiling of aquaporins during basidiocarp development in Laccaria bicolor ectomycorrhizal with Picea glauca. Mycorrhiza. 2016;26: 19–31. 10.1007/s00572-015-0643-6 [DOI] [PubMed] [Google Scholar]
- 27.Merényi Z, Prasanna AN, Zheng W, Kovacs K, Hegedus B, Bálint B, et al. Unmatched level of molecular convergence among deeply divergent complex multicellular fungi. Mol Biol Evol. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varga T, Krizsán K, Földi C, Dima B, Sánchez-García M, Sánchez-Ramírez S, et al. Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 2019;3: 668 10.1038/s41559-019-0834-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang PM, Liu XB, Dai YC, Horak E, Steffen K, Yang ZL. Phylogeny and species delimitation of Flammulina: taxonomic status of winter mushroom in East Asia and a new European species identified using an integrated approach. Mycol. Prog. 2018;17: 1013–1030. [Google Scholar]
- 30.Mizuno R, Ichinose H, Honda M, Takabatake K, Sotome I, Takai T, et al. Use of whole crop sorghums as a raw material in consolidated bioprocessing bioethanol production using Flammulina velutipes. Biosci. Biotechnol. Biochem. 2009;73: 1671–1673. 10.1271/bbb.90099 [DOI] [PubMed] [Google Scholar]
- 31.Van Peer AF, Park SY, Shin PG, Jang KY, Yoo YB, Park YJ, et al. Comparative genomics of the mating-type loci of the mushroom Flammulina velutipes reveals widespread synteny and recent inversions. PLoS One. 2011;6: e22249 10.1371/journal.pone.0022249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu XB, Li J, Yang ZL. Genetic diversity and structure of core collection of winter mushroom (Flammulina velutipes) developed by genomic SSR markers. Hereditas. 2018;155: 3 10.1186/s41065-017-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XB, Feng B, Li J, Yan C, Yang ZL. Genetic diversity and breeding history of winter mushroom (Flammulina velutipes) in China uncovered by genomic SSR markers. Gene. 2016;591: 227–235. 10.1016/j.gene.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 34.Fultz SA. Fruiting at high temperature and its genetic control in the basidiomycete Flammulina velutipes. Appl. Environ. Microbiol. 1988;54: 2460–2463. 10.1128/AEM.54.10.2460-2463.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakamoto Y, Ando A, Tamai Y, Miura K, Yajima T. Differential protein expression in the fruiting dikaryon and the non-fruiting monokaryon of Flammulina velutipes. Mycol. Res. 2001;105: 177–182. [Google Scholar]
- 36.Sakamoto Y, Akira A, Tamai Y, Miura K, Yajima T. Protein expressions during fruit body induction of Flammulina velutipes under reduced temperature. Mycol. Res. 2002;106: 222–227. [Google Scholar]
- 37.Yamada M, Sakuraba S, Shibata K, Taguchi G, Inatomi S, Okazaki M, et al. Isolation and analysis of genes specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes by fluorescence differential display. FEMS Microbiol. Lett. 2006;254: 165–172. [DOI] [PubMed] [Google Scholar]
- 38.Kim HI, Lee CS, Park YJ. Further characterization of hydrophobin genes in genome of Flammulina velutipes. Mycoscience. 2016;57: 320–325. [Google Scholar]
- 39.Liu JY, Chang MC, Meng JL, Feng CP, Zhao H, Zhang ML. Comparative proteome reveals metabolic changes during the fruiting process in Flammulina velutipes. J. Agric. Food Chem. 2017;65: 5091–5100. 10.1021/acs.jafc.7b01120 [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Lian L, Xu P, Chou T, Mukhtar I, Osakina A, et al. Advances in understanding mating type gene organization in the mushroom-forming fungus Flammulina velutipes. G3-Genes Genomes Genet. 2016;6: 3635–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33: 290 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012; 40:445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009; 37:233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S. Analysing RNA-Seq data with the DESeq package. Mol Biol. 2010; 43:1–17. [Google Scholar]
- 47.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Almási É, Sahu N, Krizsán K, Bálint B, Kovács GM, Kiss B, et al. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytol. 2019;224: 902–915. 10.1111/nph.16032 [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Zhai S, Zhang H, Zuo R, Wang J, Guo M, et al. Shared and distinct functions of two Gti1/Pac2 family proteins in growth, morphogenesis and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2014;16: 788–801. 10.1111/1462-2920.12204 [DOI] [PubMed] [Google Scholar]
- 51.Narai A, Watanabe H, Iwanaga T, Tomita T, Shimizu M. Effect of a pore-forming protein derived from Flammulina velutipes on the Caco-2 intestinal epithelial cell monolayer. Biosci. Biotechnol. Biochem. 2004; 68: 2230–2238. 10.1271/bbb.68.2230 [DOI] [PubMed] [Google Scholar]
- 52.Ota K, Leonardi A, Mikelj M, Skočaj M, Wohlschlager T, Künzler M, et al. Membrane cholesterol and sphingomyelin, and ostreolysin A are obligatory for pore-formation by a MACPF/CDC-like pore-forming protein, pleurotolysin B. Biochimie. 2013;95: 1855–1864. 10.1016/j.biochi.2013.06.012 [DOI] [PubMed] [Google Scholar]
- 53.Panevska A, Hodnik V, Skočaj M, Novak M, Modic Š, Pavlic I, et al. Pore-forming protein complexes from Pleurotus mushrooms kill western corn rootworm and Colorado potato beetle through targeting membrane ceramide phosphoethanolamine. Sci Rep. 2019;9: 5073 10.1038/s41598-019-41450-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Žurga S, Pohleven J, Renko M, Bleuler‐Martinez S, Sosnowski P, Turk D, et al. A novel β‐trefoil lectin from the parasol mushroom (Macrolepiota procera) is nematotoxic. FEBS J. 2014;281: 3489–3506. 10.1111/febs.12875 [DOI] [PubMed] [Google Scholar]
- 55.Tayyrov A, Azevedo S, Herzog R, Vogt E, Arzt S, Lüthy P, et al. Heterologous production and functional characterization of ageritin, a novel type of ribotoxin highly expressed during fruiting of the edible mushroom Agrocybe aegerita. Appl. Environ. Microbiol. 2019;85: e01549–19. 10.1128/AEM.01549-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu T, Hu C, Xie B, Zhang L, Yan S, Wang W, et al. A Single Transcription Factor (PDD1) Determines Development and Yield of Winter Mushroom (Flammulina velutipes). Appl. Environ. Microbiol. 2019;85: e01735–19. 10.1128/AEM.01735-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pelkmans JF, Lugones LG, Wösten HAB. Fruiting body formation in Basidiomycetes growth, differentiation and sexuality In: Esser Karl, editor. The Mycota I. Springer: Cham: 2016. pp. 387–405. [Google Scholar]
- 58.Luti S, Sella L, Quarantin A, Pazzagli L, Baccelli I. Twenty years of research on cerato-platanin family proteins: clues, conclusions, and unsolved issues. Fungal Biol. Rev. 2020;34: 13–24. [Google Scholar]
- 59.Gaderer R, Bonazza K, Seidl-Seiboth V. Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl Microbiol Biot. 2014;98: 4795–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamlett CA, Shirtcliffe NJ, Pyatt FB, Newton MI, McHale G, Koch K. Passive water control at the surface of a superhydrophobic lichen. Planta. 2011;234: 1267–1274. 10.1007/s00425-011-1475-z [DOI] [PubMed] [Google Scholar]
- 61.Navarro‐RóDenas A, Xu Hao, Kemppainen M, Pardo AG, Zwiazek JJ. Laccaria bicolor aquaporin LbAQP1 is required for Hartig net development in trembling aspen (Populus tremuloides). Plant Cell Environ. 2015;38: 2475–2486. 10.1111/pce.12552 [DOI] [PubMed] [Google Scholar]
- 62.Dietz S, von Bülow J, Beitz E, Nehls U. The aquaporin gene family of the ectomycorrhizal fungus Laccaria bicolor: lessons for symbiotic functions. New Phytol. 2011;190: 927–940. 10.1111/j.1469-8137.2011.03651.x [DOI] [PubMed] [Google Scholar]
- 63.Pettersson N, Filipsson C, Becit E, Brive L. Hohmann S. Aquaporins in yeasts and filamentous fungi. Biol. Cell. 2005;97: 487–500. 10.1042/BC20040144 [DOI] [PubMed] [Google Scholar]
- 64.Xu H, Cooke JEK, Zwiazek JJ. Phylogenetic analysis of fungal aquaporins provides insight into their possible role in water transport of mycorrhizal associations. Botany. 2013;91: 495–504. [Google Scholar]
- 65.Knapp DG, et al. Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep. 2018;8: 6321 10.1038/s41598-018-24686-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pellegrin C, Daguerre Y, Ruytinx J, Guinet F, Kemppainen M, Frey NF, et al. Laccaria bicolor MiSSP8 is a small-secreted protein decisive for the establishment of the ectomycorrhizal symbiosis. Environ. Microbiol. 2019;21: 3765–3779. 10.1111/1462-2920.14727 [DOI] [PubMed] [Google Scholar]
- 67.Arima T, Yamamoto M, Hirata A, Kawano S, Kamada T. The eln3 gene involved in fruiting body morphogenesis of Coprinus cinereus encodes a putative membrane protein with a general glycosyltransferase domain. Fungal Genet Biol. 2004;41: 805–812. 10.1016/j.fgb.2004.04.003 [DOI] [PubMed] [Google Scholar]
- 68.Tao Y, van Peer AF, Chen B, Chen Z, Zhu J, Deng Y, et al. Gene expression profiling reveals large regulatory switches between succeeding stipe stages in Volvariella volvacea. PLoS One. 2014;9: e97789 10.1371/journal.pone.0097789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsitsigiannis D I, Zarnowski R, Keller N P. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 2004;279: 11344–11353. 10.1074/jbc.M310840200 [DOI] [PubMed] [Google Scholar]
- 70.Ishizaki T, Shishido K. Decreased zinc ion accumulation by the basidiomycete Lentinus edodes over-expressing L. edodes priA gene. FEMS Microbiol. Lett. 2000;193: 111–5. [DOI] [PubMed] [Google Scholar]
- 71.Wang L, Tian X, Gyawali R, Upadhyay S, Foyle D, Wang G, et al. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog. 2014;10: e1004185 10.1371/journal.ppat.1004185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaur JN, Panepinto JC. Morphotype-specific effector functions of Cryptococcus neoformans PUM1. Sci Rep. 2016;6: 23638 10.1038/srep23638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L, He GJ, Chen L, Zheng J, Chen Y, Shen L, et al. Genetic basis for coordination of meiosis and sexual structure maturation in Cryptococcus neoformans. eLife. 2018;7: e38683 10.7554/eLife.38683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krüger M, Fischer R. Integrity of a Zn finger‐like domain in SamB is crucial for morphogenesis in ascomycetous fungi. EMBO J. 1998;17: 204–214. 10.1093/emboj/17.1.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukuda K, Hiraga M, Asakuma S, Arai I, Sekikawa M, Urashima T. Purification and characterization of a novel exo-beta-1,3–1,6-glucanase from the fruiting body of the edible mushroom Enoki (Flammulina velutipes). Biosci. Biotechnol. Biochem. 2008;72: 3107–3113. 10.1271/bbb.80213 [DOI] [PubMed] [Google Scholar]
- 76.Konno N, Sakamoto Y. An endo-β-1, 6-glucanase involved in Lentinula edodes fruiting body autolysis. Appl. Microbiol. Biotechnol. 2011;91: 1365 10.1007/s00253-011-3295-2 [DOI] [PubMed] [Google Scholar]
- 77.Zhou Y, Zhang W, Liu Z, Wang J, Yuan S. Purification, characterization and synergism in autolysis of a group of 1, 3-β-glucan hydrolases from the pilei of Coprinopsis cinerea fruiting bodies. Microbiology. 2015;161: 1978–1989. 10.1099/mic.0.000143 [DOI] [PubMed] [Google Scholar]
- 78.Chae E, Tan QKG, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135: 1235–1245. 10.1242/dev.015842 [DOI] [PubMed] [Google Scholar]
- 79.Xu G, Ma H, Nei M, Kong H. Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. U. S. A. 2009;106: 835–840. 10.1073/pnas.0812043106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190: 1157–1195. 10.1534/genetics.111.128033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gowda NKC, Kandasamy G, Froehlich MS, Dohmen RJ, Andréasson C. Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc. Natl. Acad. Sci. U. S. A. 2013;110: 5975–5980. 10.1073/pnas.1216778110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X, Ren A, Li MJ, Cao PF, Chen TX, Zhang G, et al. Heat stress modulates mycelium growth, heat shock protein expression, ganoderic acid biosynthesis, and hyphal branching of Ganoderma lucidum via cytosolic Ca2+. Appl. Environ. Microbiol. 2016;282: 4112–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang GZ, Ma CJ, Luo Y, Zhou SS, Zhou Y, Ma XL, et al. Proteome and transcriptome reveal involvement of heat shock proteins and indoleacetic acid metabolism process in Lentinula edodes thermotolerance. Cell. Physiol. Biochem. 2018;50: 1617–1637. 10.1159/000494784 [DOI] [PubMed] [Google Scholar]
- 84.van der Laan R, Roest HP, Hoogerbrugge JW, Smit EM, Slater R, Baarends WM, et al. Characterization of mRAD18Sc, a mouse homolog of the yeast postreplication repair gene RAD18. Genomics. 2000;69: 86–94. 10.1006/geno.2000.6220 [DOI] [PubMed] [Google Scholar]
- 85.Verkade H, Teli T, Laursen L, Murray J, O’Connell M. A homologue of the Rad18 postreplication repair gene is required for DNA damage responses throughout the fission yeast cell cycle. Mol. Genet. Genomics. 2001; 265:993–1003. 10.1007/s004380100494 [DOI] [PubMed] [Google Scholar]
- 86.de Padula M, Slezak G, Auffret van Der Kemp P, Boiteux S. 2004. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7, 8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32: 5003–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]