Figure 4.
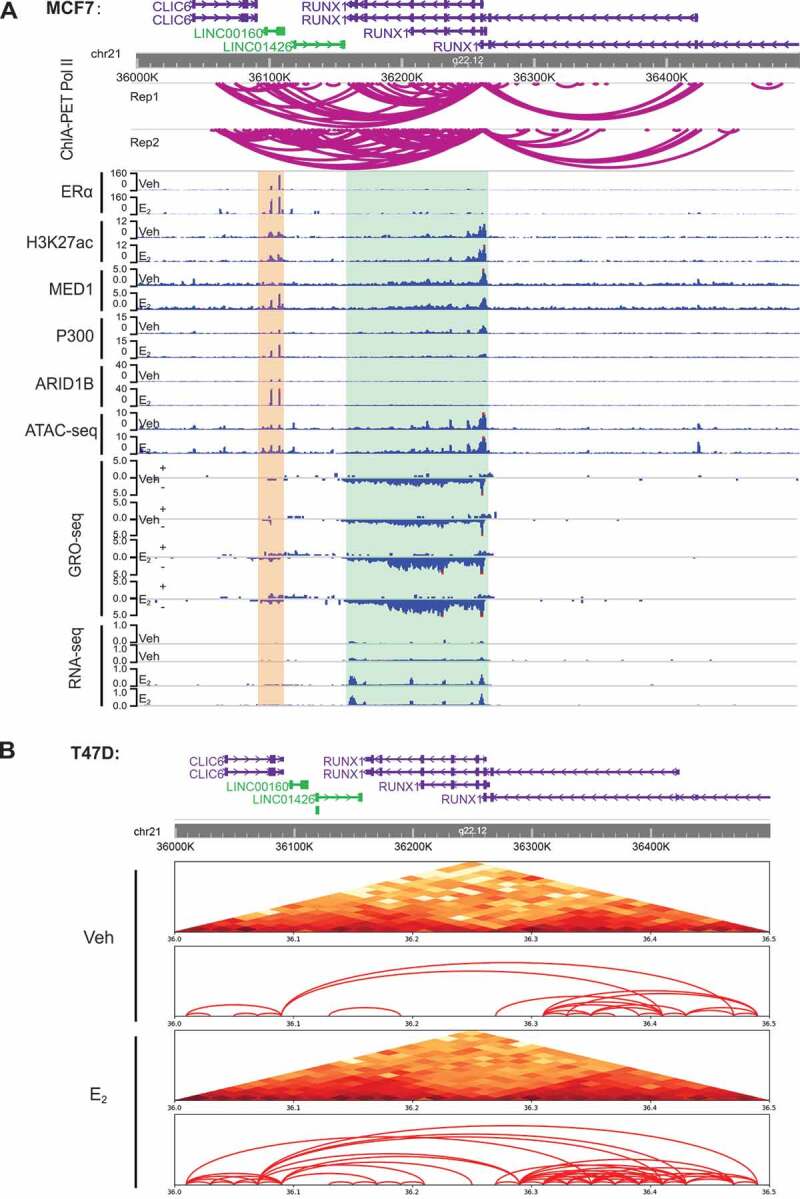
LINC00160 and RUNX1 are co-regulated by E2 and are located within one TAD.
A. The genome browser views of Pol II ChIA-PET signals and various epigenomics-based sequencing data for the chromatin region that contains both LINC00160 and RUNX1. Top: Schematic diagram showing the promoter-enhancer interactions between regions around the LINC00160and RUNX1 measured by Pol II ChIA-PET (n = 2, biological replicates).Bottom: The ChIP-seq data of ERα, Η3Κ27ac, P300, MED1, ARID1B as well as the ATAC-seq, GRO-seq and RNA-seq data around the LINC00160 (marked with yellow shadow) and RUNX1 (marked with green shadow) loci in MCF7 cell line. The results demonstrate a coordinated E2-induction for both LINC00160 and RUNX1 andstrong interactions between the RUNX1 promoter and several ERα enhancers located inside LINC00160 gene. B. Heatmap (top) and schematic diagram (bottom) representing chromatin interactions within 500kb regions covering LINC00160 and RUNX1 measured by Hi-C experiments in the T47D cell line (+- E2). The Hi-C results suggest that oestrogen enhances the promoter-enhancer interactions between LINC00160 and RUNX1 and these two genes are regulated as a 3D structural unit supported by a TAD domain that containing both LINC00160 and RUNX1 loci.
