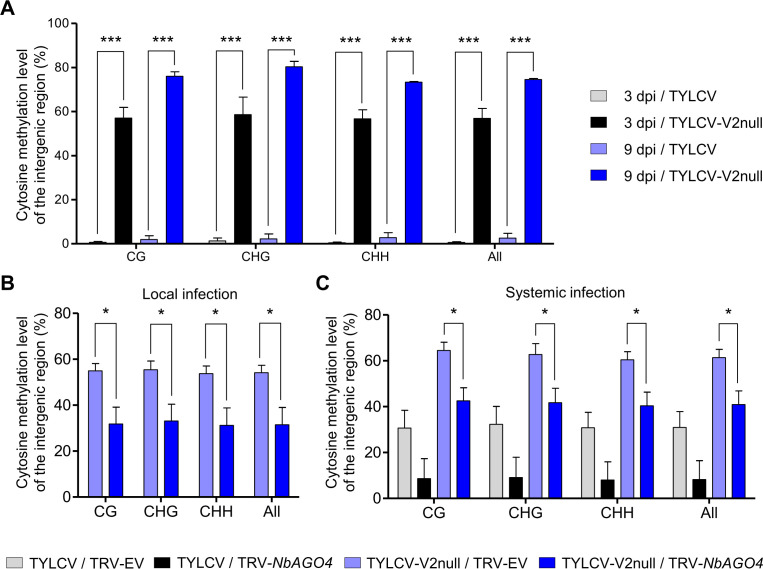
Figure 4. V2 suppresses the AGO4-dependent methylation of viral DNA.
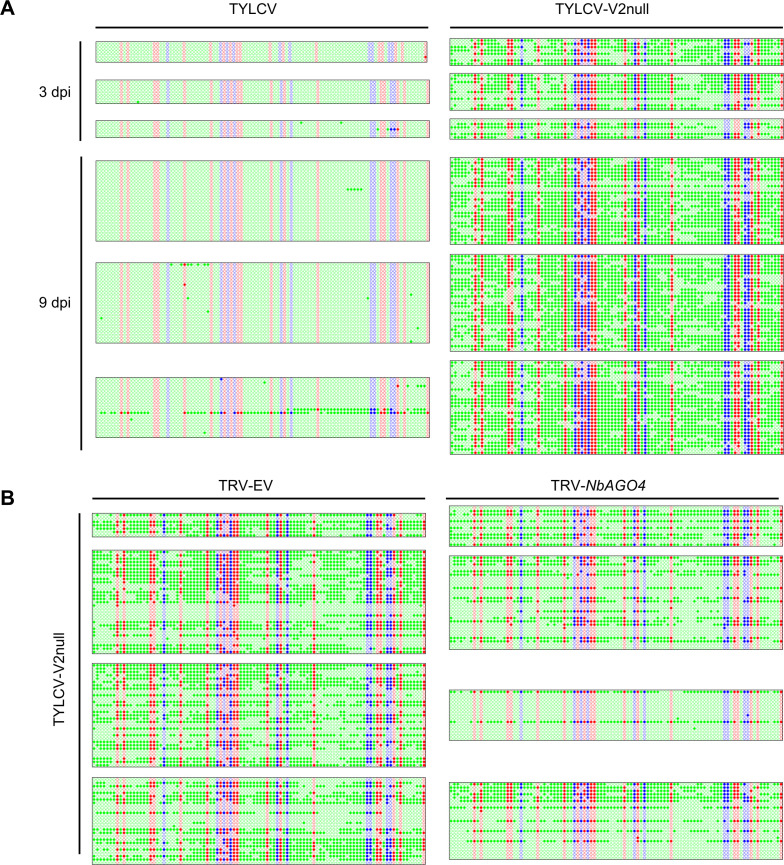
(A) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in local infection assays with TYLCV wild-type or V2 null mutant (TYLCV-V2null) in N. benthamiana at 3 or 9 days post-inoculation (dpi), as detected by bisulfite sequencing. The original single-base resolution bisulfite sequencing data are shown in Figure 4—figure supplement 1A. Values are the mean of three independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. ***: p<0.001. The values of cytosine methylation in each biological replicate are shown in Supplementary file 1. (B) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in local infection assays with the V2 null mutant TYLCV (TYLCV-V2null) in AGO4-silenced (TRV-NbAGO4) or control (TRV-EV) N. benthamiana plants at 4 dpi, as detected by bisulfite sequencing. Samples come from the same plants used in Figure 3E. The original single-base resolution bisulfite sequencing data are shown in Figure 4—figure supplement 1B. Values are the mean of four independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05. The values of cytosine methylation in each biological replicate are shown in Supplementary file 1. (C) Percentage of methylated cytosines in the intergenic region (IR) of TYLCV in systemic infection assays with TYLCV wild-type or V2 null mutant (TYLCV-V2null) in AGO4-silenced (TRV-NbAGO4) or control (TRV-EV) N. benthamiana plants at 3 weeks post-inoculation (wpi), as detected by bisulfite sequencing. Samples come from the same plants used in Figure 3F. The original single-base resolution bisulfite sequencing data are shown in Figure 4—figure supplement 2. Values are the mean of four independent biological replicates; error bars indicate SEM. Asterisks indicate a statistically significant difference according to Student’s t-test. *: p<0.05. The values of cytosine methylation in each biological replicate are shown in Supplementary file 1.