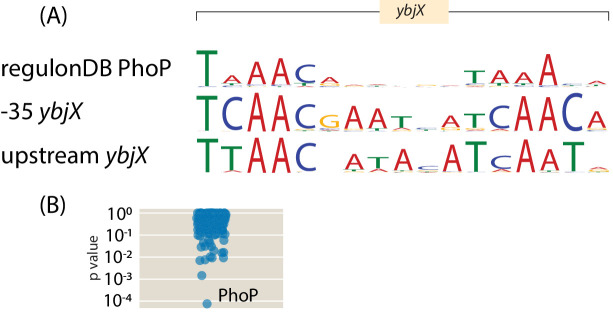
Appendix 3—figure 2. Motif comparison using TOMTOM for the two PhoP-binding sites in the ybjX promoter.
Searching our energy motifs against the RegulonDB database using TOMTOM allowed us to guide our transcription factor knockout experiments. Here, we show the sequence logos of the PhoP transcription factor from RegulonDB (top) and the ones generated from the ybjX promoter energy matrix. E-value = 0.01 using Euclidean distance as a similarity matrix.