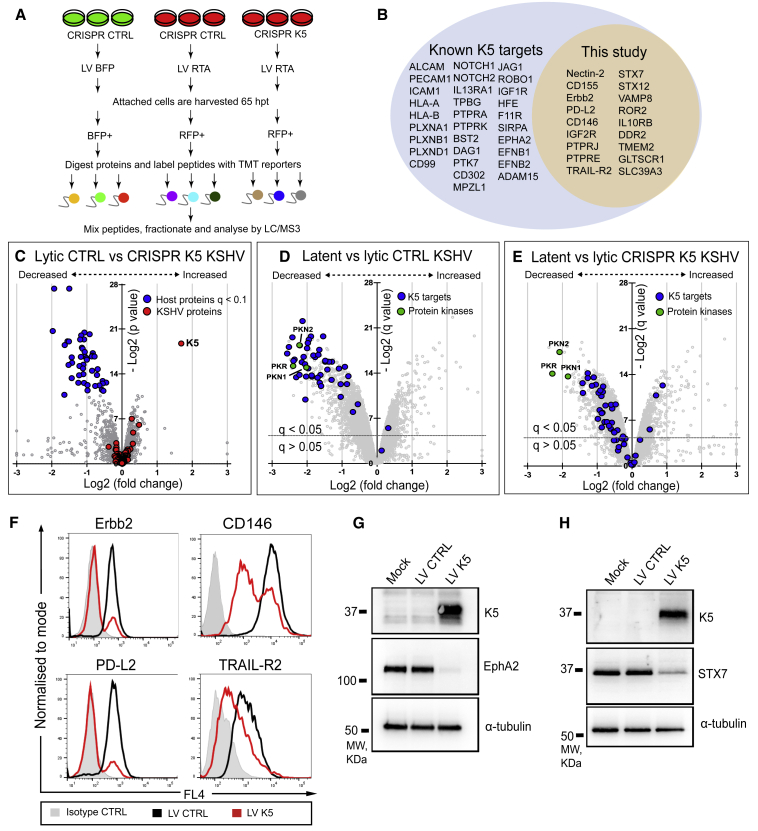
Figure 3.
CRISPR Proteomics Identifies 48 KSHV K5 Targets in Endothelial Cells
(A) Schematic overview of the quantitative proteomic analysis of cells with lytic versus latent KSHV infection. HuAR2T.rKSHV.219 Cas9 cells harboring control or K5-specific sgRNAs were transduced with LV RTA or control LV BFP, sorted on BFP+ or RFP+, and analyzed by MS.
(B) KSHV K5 protein significantly (q < 0.1) downregulates 48 proteins in endothelial cells, of which 30 are known K5 targets (left side of blue circle) and 18 proteins represent K5 targets identified in this study (yellow circle).
(C–E) Scatterplots display pairwise comparison between lytic control and CRISPR K5 (C), latent and lytic control (D), and latent and lytic CRISPR K5 (E) KSHV infections. Each point represents a single protein, plotted by its log2 (fold change in abundance) versus the statistical significance (p and q value) of that change. Values were corrected for multiple hypothesis testing using the method of Benjamini-Hochberg. Dotted lines: q = 0.05.
(F) Flow cytometry analysis of HuAR2T cells transduced with LV K5 or control LV and stained with isotype control antibody or antibody specific for the indicated proteins.
(G and H) Immunoblot analysis of HuAR2T cells transduced with LV K5 or control LV, sorted on GFP+, and probed with the indicated antibody.