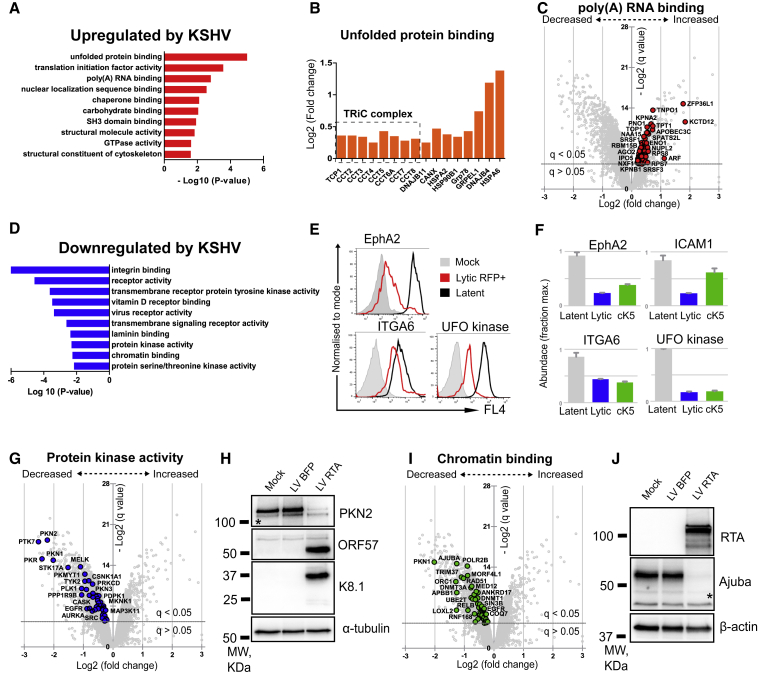
Figure 5.
DAVID GO Term Analysis of Proteins Dysregulated by Lytic KSHV Infection
(A) Ten most enriched GO terms ranked by statistical significance (p value) in the category “molecular function” among proteins upregulated by lytic KSHV.
(B) Histogram shows the fold change in the abundance of the proteins from the GO term “unfolded protein binding.”
(C) Upregulated proteins (red points) from the GO term “poly(A) RNA binding” are highlighted on the scatterplot that displays pairwise comparison between latent and lytic KSHV infections. Each point represents a single protein, plotted by its log2 (fold change in abundance) versus statistical significance (q value) of that change. Value was corrected for multiple hypothesis testing using the method of Benjamini-Hochberg. Dotted line: q = 0.05
(D) Ten most enriched GO terms ranked by statistical significance (p value) in the category “molecular function” among proteins downregulated by lytic KSHV.
(E) Validation of lytic KSHV-mediated downregulation of the surface proteins from the GO terms “integrin binding” and “transmembrane receptor protein tyrosine kinase activity.”
(F) Relative abundance of the indicated host proteins in the cells with latent (gray), lytic CRISPR control (blue) and lytic CRISPR K5 (green) KSHV infections. Protein abundance is calculated as a fraction of the maximum TMT reporter ion intensity. Data are represented as mean ± SEM.
(G–J) Downregulated proteins from the GO terms “protein kinase activity” (G) and “chromatin binding” (I) are highlighted on the scatterplots that display pairwise comparison between latent and lytic KSHV infection. Each point represents a single protein, plotted by its log2 (fold change in abundance) versus statistical significance (q value) of that change. Values were corrected for multiple hypothesis testing using the method of Benjamini-Hochberg. Dotted line: q = 0.05. (H and J) Immunoblot analysis of HuAR2T.rKSHV.219 cells transduced with LV RTA or control LV BFP, sorted on BFP+ or RFP+, and probed with the indicated antibody. Non-specific bands are labeled with an asterisk.