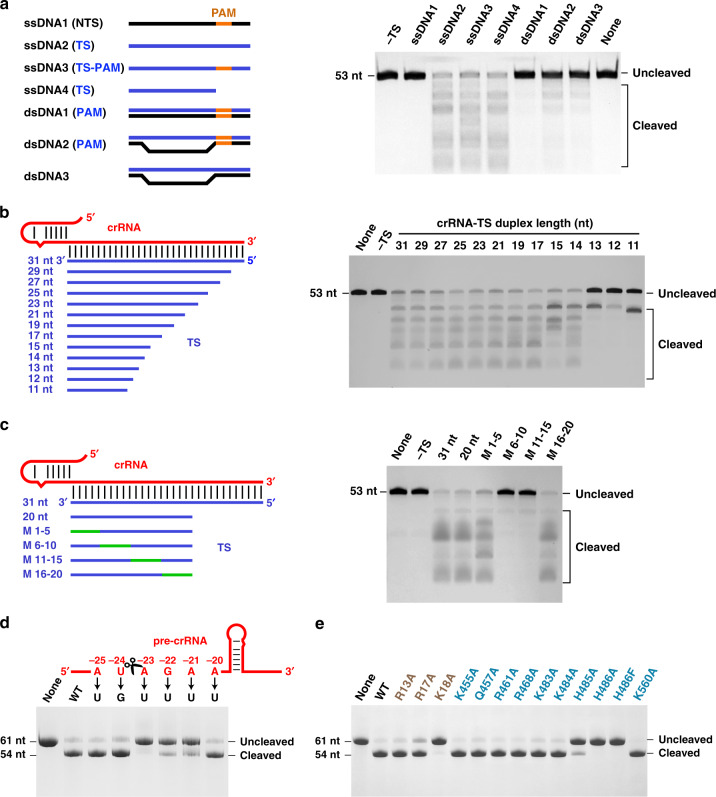
Fig. 6. Cas12i2 target recognition activates non-specific ssDNA cleavage and pre-crRNA processing by Cas12i2.
a The formation of the guide:target heteroduplex activates the RuvC catalytic pocket, inducing the trans cleavage of ssDNA in a PAM-independent manner. (Left) Schematic representation of ssDNA and dsDNA activators. The complementary target DNA and non-complementary DNA are in blue and black, respectively. The PAM sequence is highlighted in orange. (Right) The DNA cleavage assay using the 200 nM 5′-Cy3-labeled ssDNA substrate in the presence of 200 nM DNA activators at 37 °C for 15 min. b The length of the complementary ssDNA activator affects non-specific DNA cleavage. (Left) Schematic representation of crRNA and ssDNA activators that is complementary to the guide RNA. (Right) The ssDNA cleavage assay in the presence of ssDNA activators with various lengths. c Mismatches between the target strand DNA and the crRNA spacer region affect activation of the RuvC catalytic pocket. (Left) Schematic representation of crRNA and ssDNA with mismatched regions are highlighted in green. (Right) The ssDNA cleavage assay in the presence of ssDNA activators containing mismatches. d Denaturing gel analysis showing that pre-crRNA processing by Cas12i2 depends on the crRNA repeat sequence shown in the up panel directly downstream of the cleavage site. The cleavage site is indicated by cartoon scissors. The pre-crRNA cleavage assay using the pre-crRNA mutants substrate in the presence of Cas12i2 at 37 °C for 30 min. e Pre-crRNA processing using Cas12i2 mutants.