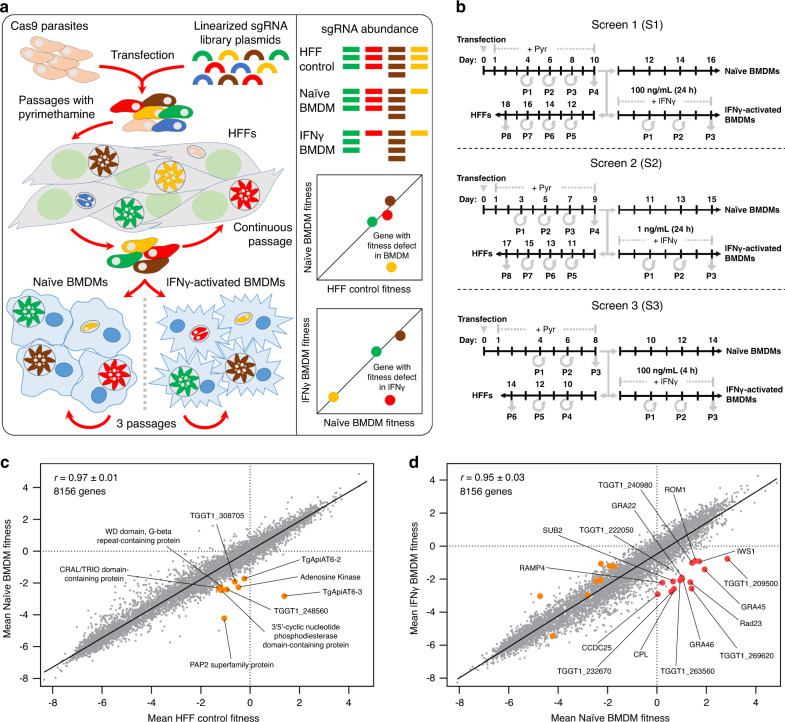
Fig. 1. Toxoplasma gondii genome-wide loss-of-function screen in naïve or IFNγ-activated murine bone-marrow-derived macrophages.
a Screening workflow. At least 5 × 108 Cas9-expressing RH parasites were transfected with linearized plasmids containing 10 sgRNAs against every Toxoplasma gene. Transfected parasites were passaged in HFFs under pyrimethamine selection to remove non-transfected parasites and parasites that integrated plasmids with sgRNAs targeting parasite genes important for fitness in HFFs. Subsequently, the pool of mutant parasites was either continuously passaged in HFFs or passaged for three rounds in murine BMDMs that were left unstimulated or prestimulated with IFNγ. The sgRNA abundance at different passages, determined by illumina sequencing, was used for calculating fitness scores and identifying genes that confer fitness in naïve or IFNγ-activated BMDMs. b Timeline for the generation of mutant populations and subsequent selection in the presence or absence of murine IFNγ. Times at which parasites were passaged (P) are indicated. c Correlation between mean parasite gene fitness scores in Naïve BMDMs and HFF control. Orange dots indicate nine high-confidence candidate genes that confer fitness in naïve BMDMs compared to HFFs (Table 1). d Correlation between mean parasite gene fitness scores in IFNγ-activated and Naïve BMDMs. Red dots indicate 16 high-confidence IFNγ fitness-conferring candidate genes in murine BMDMs and orange dots the nine high-confidence genes conferring fitness in naïve BMDMs compared to HFFs (Table 1).