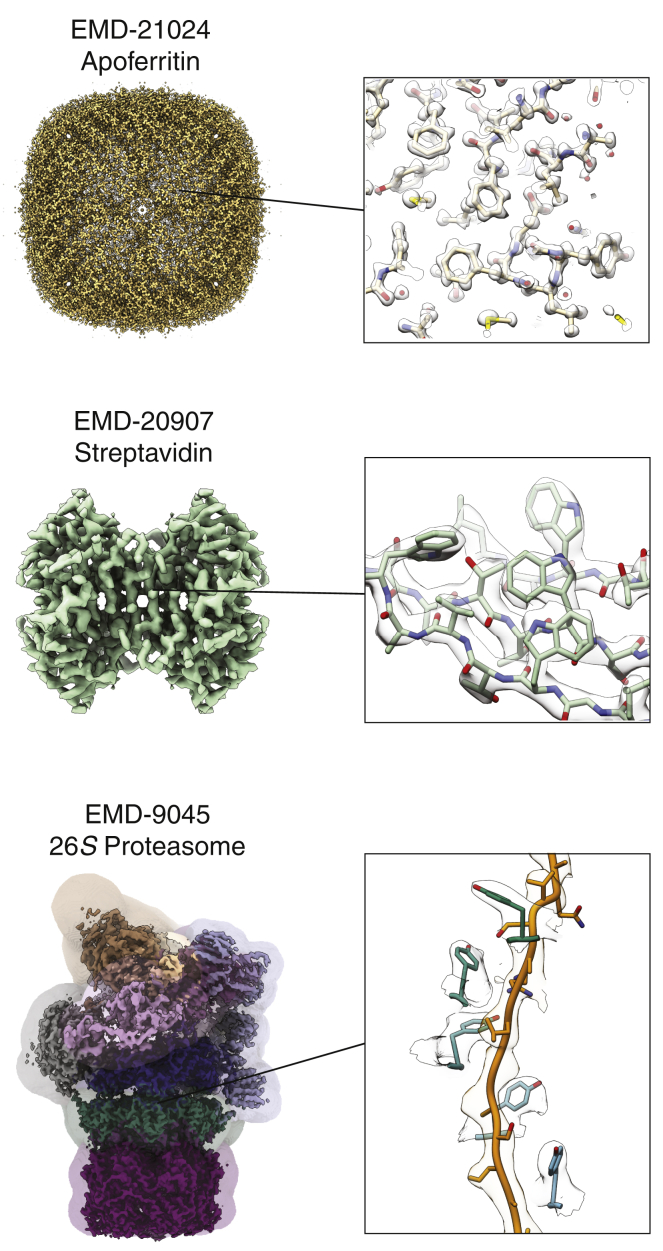
Figure 2.
Cryo-EM structures of biological macromolecules enabled by technical and methodological advances in SPA. Top: shown is mouse apoferritin determined to ∼1.2-Å resolution using a TEM equipped with a cold FEG and next-generation energy filter and direct detector. Middle: shown is the ∼2.6-Å resolution reconstruction of the 52-kDa streptavidin imaged over graphene monolayer support grids. Bottom: shown is the ∼4.2-Å resolution reconstruction of the substrate-engaged yeast 26S proteasome in the "4D" motor state. Focused classification and refinement strategies (colored by focusing area) were used to identify the distinct motor conformations and to generate composite reconstructions to facilitate model building for each state. The inset on the right shows density features of the substrate polypeptide encircled by a spiral-staircase arrangement of pore loop tyrosine residues within the central pore of the AAA+ motor. The EMDB accession code and full map (left) and density features with the corresponding atomic model docked in (right) are shown for each structure. To see this figure in color, go online.