Fig. 5.
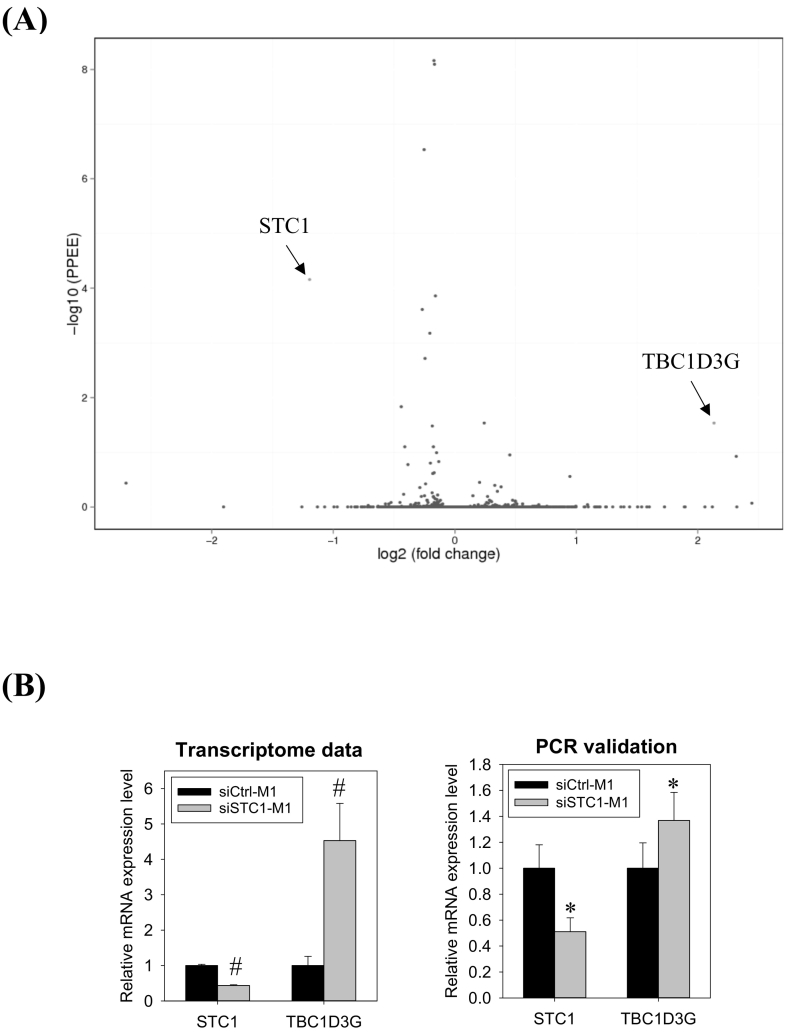
Transcriptomic analysis of STC1 knockdown M1 cells. (A) Volcano plot of differentially expressed genes (DEGs). The x-axis represents the differential gene expression (log2). Y-axis represents the p-value (−log10). The red dots represent the significantly up-regulated DEG - TBC1D3G and the down-regulated DEG - STC1. (B) Messenger RNA levels of STC1 and TBC1D3G, as revealed by transcriptome (the left panel) and validated by real-PCR (the right panel). Data were presented as the mean ± S.D. Pounds (#) indicate statistical significance relative to the respective control (posterior probability of equal expression, PPEE, Supplementary file 1). Asterisks denote statistical significance relative to the respective control (*p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Transcriptomic analysis of STC1 knockdown M1 cells. (A) Volcano plot of differentially expressed genes (DEGs). The x-axis represents the differential gene expression (log2). Y-axis represents the p-value (−log10). The red dots represent the significantly up-regulated DEG - TBC1D3G and the down-regulated DEG - STC1. (B) Messenger RNA levels of STC1 and TBC1D3G, as revealed by transcriptome (the left panel) and validated by real-PCR (the right panel). Data were presented as the mean ± S.D. Pounds (#) indicate statistical significance relative to the respective control (posterior probability of equal expression, PPEE, Supplementary file 1). Asterisks denote statistical significance relative to the respective control (*p < 0.05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)