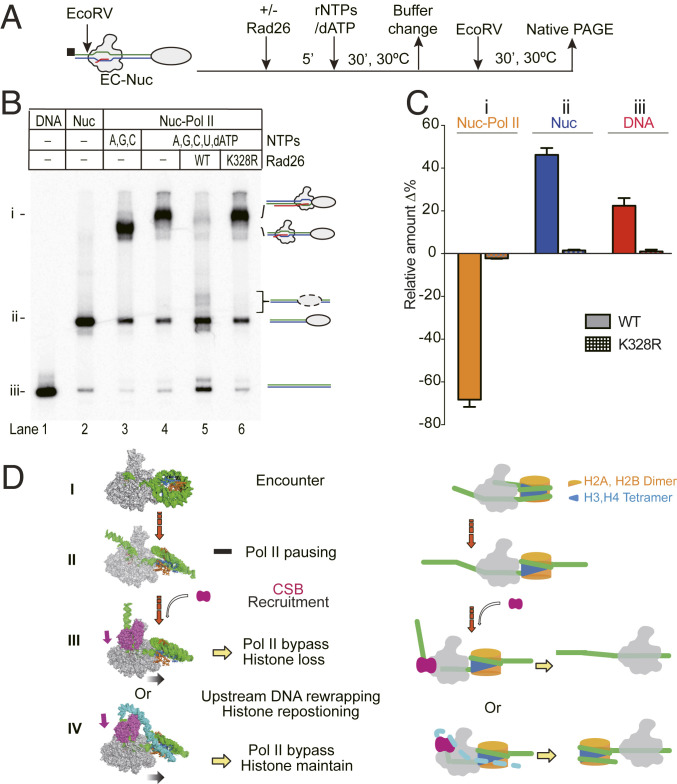
Fig. 5.
Fate of the nucleosome after Rad26-mediated Pol II transcription bypass. (A) Experimental approach for analyzing the fate of nucleosomes after Rad26-mediated Pol II transcription bypass. The biotin label is shown as a black square. An EcoRV cleavage site was introduced in the upstream DNA to release the DNA from biotin beads. (B) Analysis of the released product by native polyacrylamide gel electrophoresis (PAGE). Three different species, the Pol II-Nuc complex, Nuc only, and DNA only, are labeled as i, ii, and iii, respectively, on the left. The mobilities of the released product are indicated on the right. In the presence of active Rad26, the majority of the arrested Pol II-nucleosome complex was converted into nucleosome bands. (C) Quantitation of the native PAGE assay. Lanes 4–6 from native gel were compared. The experiments were performed three times and shown as means with error bars. (D) Scheme of Rad26-mediated Pol II transcription bypass of a nucleosome. Pol II transcription encounters core nucleosome at SHL-6 (step I) and, then, proceeds to the major pausing site at SHL-1 (step II). In this step, an H2A-H2B dimer is exposed. Pausing of Pol II results in the recruitment of Rad26 which binds to the upstream of DNA of the elongation complex. Rad26 promotes the forward translocation of Pol II and helps Pol II bypass the nucleosome barrier (step III). Rad26 may also rewrap the upstream DNA and cover the exposed H2A-H2B to prevent the dissociation of the H2A-H2B dimer (step IV).