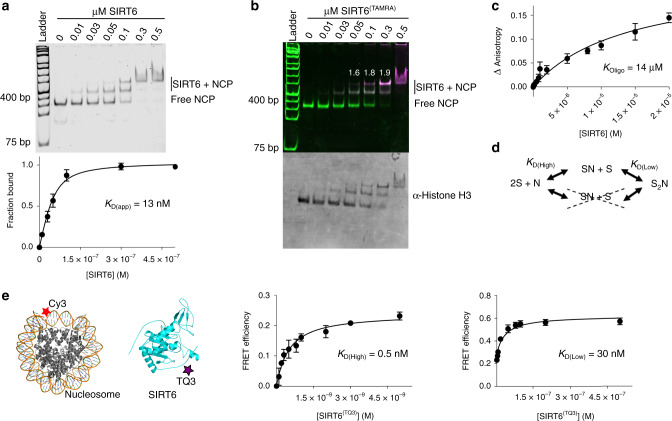
Fig. 1. SIRT6 and NCPs assemble with low nanomolar affinity via a two-site binding mechanism.
a EMSA of NCPs bound to varying SIRT6 concentrations, then detected by SYBR Safe fluorescence. Data are presented as the mean ± s.d. from six independent experiments. The apparent KD was calculated using a ligand-depleted equation. b SIRT6:NCP stoichiometry was determined by detecting fluorescence from both SYBR Safe-stained DNA and TAMRA-labeled SIRT6. The white numbers indicate the relative TAMRA signal in the higher shifted band compared to the lower bound band, both normalized to DNA content. The gel was subsequently blotted for histone H3. The image is representative of three independent experiments. c To measure SIRT6 self-oligomerization, the fluorescence anisotropy of pyrene-labeled SIRT6 was monitored with titration of unlabeled SIRT6. Data are presented as mean ± s.d. from three independent experiments. d The data indicates a two-site binding mechanism, in which two SIRT6 molecules bind at separate sites on a nucleosome. e A FRET assay was employed to measure binding between 1 nM of Cy3-labeled nucleosomes and SIRT6 labeled with TQ3, which accepts energy transfer from Cy3. The titration reveals a two-site binding curve (Supplementary Fig. 2f). The intermediate binding constant KD(High) was fitted to a ligand-depleted equation from titration points up to 5 nM SIRT6(TQ3), while KD(Low) was fitted to a specific-binding equation from data points ranging from 5 nM to 500 nM. Data are presented as mean ± s.d. from four independent experiments. Source data are provided as a source data file.