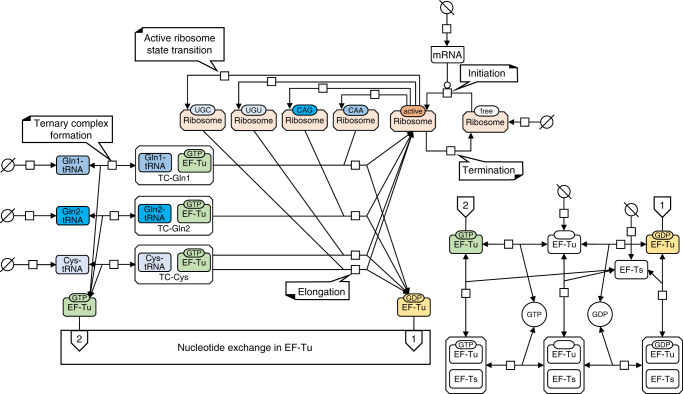
Fig. 1. Schematic overview of the translation model.
A reduced pathway for elongation with amino acids cysteine (aminoacyl-tRNA Cys) and glutamine (aa-tRNA Gln1, Gln2) is represented in Systems Biology Graphical Notation. Initiation: free (unbound) ribosome gets converted to active ribosome, modulated by mRNA. Termination: active ribosome converts back to free ribosome at a rate fixed by the desired protein production rate. Active ribosome state transition: active ribosome instantaneously binds to codons (61 codons in full model, 4 here) at the fractions set by the specified proteome composition. Ternary complex formation: charged tRNAs (40 aa-tRNAs in full model, 3 here), replenished from a pool, combine with EF-Tu*GTP to form ternary complexes (40 TCs in the full model). Kinetic parameters of these reversible processes depend on the aa-tRNA. Elongation: labeled ribosome binds with the cognate TC to elongate the protein with the respective amino acid. The ribosome returns to its active state and EF-Tu*GDP is released. Other products of this reaction, such as deacylated tRNA, are not modeled. Nucleotide exchange (see right panel): EF-Tu*GDP is reactivated to EF-Tu*GTP in a sequence of steps modeled by reversible mass action kinetics. GTP and GDP pools are modeled with fixed concentrations. The nucleotide exchange is supported by EF-Ts, and the main flux is carried through the complexes formed by EF-Tu with EF-Ts.