Abstract
Background
The aim of the present study was to investigate the effect of over-expressing circular RNA (circ_0003645) on cell functions and its molecular mechanism in breast cancer.
Methods
The expression profile of circ_0003645, breast cancer cell lines, and the transcription levels of circular RNA, miRNA and HMGB1 gene were detected by qRT-PCR. Flow cytometry analysis was manipulated to evaluate cancer cell proliferation and cell apoptosis. The correlation between miR-139p-3p and circular_0003645 or HMGB1 was predicted by GEO, and TCGA was confirmed using the dual-luciferase reporter assay.
Results
Circ_0003645 expression was conspicuously increased in both the breast cancer tissues and cell lines. Circ_0003645 knockdown inhibited cell proliferation and induced the apoptosis of breast cancer cells in vitro and in vivo. By sponging miR-139-3p, circ_0003645 promoted the breast cancer cells progression and positively regulated HMGB1 gene.
Conclusion
Circ_0003645 functions as a ceRNA for miR-139-3p, which could upregulate HMGB1 and further promote cell proliferation in breast cancer.
Keywords: circular RNA, cancer pathogenesis target, knockdown, miRNA, proliferation
Introduction
Worldwide, breast cancer is the most common cancer affecting women and ranks as the second highest cause of cancer-related death in women.1,2 Despite intensive investigation and therapeutic improvement, there are few specific biomarkers and this restricts the early diagnosis of breast cancer.3,4 It is well known that early diagnosis of breast cancer is an effective method to reduce the morbidity and mortality, and this has become one of the primary challenges in the treatment of this disease.5,6
As a family of naturally endogenous noncoding RNAs (ncRNAs), circular RNAs (circRNAs) contain structures distinct from other ncRNAs, constituting a class of 3ʹ-to-5ʹ covalently closed and cyclized RNAs derived through either natural spliceosome mediation or lariat structure such as an upstream splice acceptor ligating a downstream splice donor.7,8 Thus circRNAs lack 5ʹ caps or 3ʹ tails, most of them have no protein coding capability and only a small part can be translated into polypeptides.9–11 Owing to these special structures, circRNAs exhibit resistance to RNase R, which ensures their species conservation.12,13 In addition, circRNAs contain miRNA response elements (MREs). The MREs, an important part of RNA, which can play a key role in gene regulation by competing for miRNA, allow researchers to sequester RNA binding protein (RBPs, a critical regulator of RNA metabolism) and find miRNA-specific antagonists.14–16 More and more evidence demonstrates that circRNAs may serve as miRNA sponges to regulate gene expression.17 Recent studies also suggested that circRNAs were widespread and critical to gene expression in mammals18–20 and highly prevalent in human eukaryotic transcriptomes.
Discovery of the underlying pathophysiological mechanisms through study of circular RNAs in tumor patients could improve the diagnosis and therapeutics of these cancers. Developments in the field of circRNA concerning breast cancer are recent. With more and more patients suffering from this cancer there is an urgent need for new biomarkers and treatment targets. In a previous study, a novel circular RNA (circ_0003645) was highly expressed in lung cancer cells, endothelial cells and hepatocellular carcinoma cells and regulated cell progression through the ceRNA mechanism.21 Nevertheless, the expression profiles and functions of circ_0003645 in breast cancer remain largely unknown. Other studies reported that miR-139-3p showed relatively lower expression in malignant glioma, liver cancer and cervical cancer cases, and it could inhibit the proliferation, migration and metastasis of tumor cells.22–25
In this study, we demonstrated the newly detected roles of circRNAs by verifying upregulation of circ_0003645 and its oncogenic function in breast cancer cells. Mechanistically, we found that the miR-139-3p pathway was partially responsible for its oncogenic role in the disease. Collectively, the present study documented a novel circRNA in the progression of breast cancer. Thus, we argue that circ_0003645 can become an essential element of the diagnostic and therapeutic targets for breast cancer in the future.
Materials and Methods
Patient Enrollment and Tissues Collection
Forty-five pairs of breast tumor tissues and adjacent breast tissues were obtained from patients in the Department of Breast and Thyroid Surgery from TongJi Hospital. The study was approved by the ethical committee of the Affiliated Hospital of TongJi Hospital. Each patient signed informed consent and permitted tissue extraction. Collection of breast cancer tissues undergone in surgery were from the Department of Emergency Medicine, Tongji Hospital of Tongji Medical College. Non-tumorous tissues were acquired at sites 10 cm away from the pathological tissues. The stages and classification of collected tumors were histologically verified by corresponding pathologists. Breast cancer cell lines, consisting of T47D, MCF-7, BT549, MDA-MB-231 and Hs-578T, respectively, together with normal breast epithelial cell lines (MCF-10A and MCF-12A) were purchased and obtained from the Shanghai Institute of Chinese Academy of Sciences Cell Collection.
Bioinformatic Identification of circRNAs
Using the Gene Expression Omnibus database (GEO), we compared 8 breast tumor tissues and 3 corresponding tissues from the expression profiles of hard-clipped reads and unmapped reads. Among the 8 breast cancer samples, there were two different molecular subtypes, luminal A (LA) and triple-negative breast cancer (TNBC).26 By searching circRNA and breast cancer key words, we found the required data types according to series, platform, samples, species and history. Then, we downloaded these data from ftp server. With this information, we used Entrez system in NCBI homepage to search circRNAs materials related with breast cancer. Later, by using data analysis tools, we obtained gene expression profiles for circular RNAs information in breast cancer datasets. Using GEO2R software (the function of the tool is to import the data in the GEO database into the R language, and then perform the difference analysis) was to compare several sets of groups of samples in GEO series and to identify genes that were differentially expressed circular RNA across experimental conditions.27 We predicted the binding site of miR-139-3p within cir_0003645 by using the CircInteractome database (https://circinteractome.nia.nih.gov/) following standard procedures and predicted the miRNA-binding sites of cir_0003645 and HMGB1 using TargetScan (http://www.targetscan.org/).28 The Cancer Genome Atlas Program (TCGA) was available for discovering miR-139-3p expression in breast cancer cases and the intuitive data figure made by Starbase portal.29
Nuclear/Cytoplasmic Fractionation and Quantitative RT-PCR
NR-PER nuclear and cytoplasmic extraction reagent (catalog no. 78833; Pierce) were available for extraction of the nuclear and cytoplasmic RNA from tumor samples using 1×107 breast cancer cells, according to the manufacturer’s instructions in the presence of RiboLock RNase inhibitor (catalog no. EO0381; Thermo Scientific), while isolation of total RNA from whole-cell lysates with Rnase R treatment was achieved with RNAesay mini kit (Qiagen Valencia, CA USA). For the detailed experiment information described, 750 L TRIzol reagent was instilled into 250 L of each fraction for RNA extraction. Total RNA equivalent 1g volume from each fraction was used for cDNA synthesis, and RT-PCR analysis results provided the expression level of the indicated circRNAs in each fraction. To evaluate the quality of the fractionation assay to control immuno-blotting, nuclear marker was available from ORC-2 (catalog no. 551,178; BD Biosciences), and cytoplasmic marker was LAMP-1.
RNase R treatment and quantitative RT-PCR procedures were the following: incubation of 1 μg volume of DNAase-digested RNA for 60 min at 37°C with or without 3 U μg−1 of RNase R (Epicentre Technologies, Madison, WI), subsequently purifying the resulting RNA using an RNeasy MinElute cleaning Kit (Qiagen, Valencia, CA, USA), performing cDNA synthesis from 1 μg total RNA in 20 μL reaction volume using Bio-rad select cDNA synthesis kit with random primer for circRNA experiments, then using TaqMan universal master mix II with uracil-N-glycosylase (UNG) (catalog no. 44,400; Applied Biosystems) for quantitative PCR (qPCR) analysis of cDNA samples, achieving direct quantification of RNA derived from serum and plasma with an iTaq universal SYBR green one-step kit (catalog no. 172–515; Bio-Rad). In the qPCR analysis, GAPDH is used as a housekeeping gene in the cytoplasm and U6 as a housekeeping gene in the nucleus. The process of qPCR is to pre-denature at 95°C for 1min, then denature for 10 s, lower the temperature to 60°C for 20 s, and then raise to 72°C for 10 s, repeat the process 35 times, and finally, keep the temperature at 72°C for 2 min. After the program is completed, use the 2−ΔΔCT formula to calculate the relative expression.
Transfections and Knockdown
Lentivirus with circ_0003645 over-expression vector was transfected into two cell lines (MCF-7 cells and MDA-MB-231) by adding puromycin into the stable cells. Circular RNA knock-down procedure was using validated Stealth RNAi siRNA against circ_0003645 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Briefly, seed 4 × 105 cells in 60 mm plate the day before si-RNA treatment. Then, on the day of operation, mix 20 nM transfection solution with HiPerFect and serum free media according to protocol suggested amount to produce a final volume of 100 μL transfection reagent mixture. Incubate the mixture solution for 15 min to form transfection complexes. Instill 100 μL solution by drips into the cells and keep the mixture in an incubator and monitor the gene silencing.
Luciferase Reporter Assay
Plasmid of circ_0003645 were co-infected with miR-139-3p into breast cancer cells seeded in a 96-well plate. The miR-NC was used as negative control. The cells were collected after 48 h, and the Firefly and Renilla luciferase activities were determined with a dual-luciferase reporter assay system. The relative luciferase activity was normalized to Renilla luciferase activity.
Cell Proliferation Assay
Detection of cell proliferation was done with a CCK-8 assay kit by culturing cells at a density of 3 × 103 per well in a 96-well plate in DMEM supplemented with 10% FBS and anti-mycotic antibiotic. Different experimental time points were set as following: 0, 24, 48, 72 and 96 hours. 10 µ L of CCK-8 reagent (CellTiter 96® aqueous one solution reagent, Promega, Madison, WI) was directly added into the culture medium for the indicated time according to the manufacturer’s instructions. For determination of cell proliferation, procedures from CellTiter-Glo assay (Promega Madison USA) were followed: CellTiterGlo reagent was added directly into the cells in 96-well plates and the OD value measured with the luminescence at 450 nm after incubation for 2.5 h at 37°C on an Envision reader (PerkinElmer).
Colony Formation Assay
MCF-7 and MDA-MB-231 cells (1×103 cells/well) were seeded in 6-well plates and the plates incubated at 37°C for 7 days, transferring medium every 2 days. On day 7, cells were fixed with paraformaldehyde (4%) and stained with a crystal violet solution. The cell colonies numbers were counted and the results analyzed.
Flow Cytometric Analysis
To investigate cell apoptosis, we washed digested breast cancer cells with cold PBS and re-suspended cells in binding buffer to fix them. 5 µL Annexin-APC was added into cell solution. After mixing the solution, 5 µL 7-ADD staining solution was added, then they were incubated at room temperature in the dark for 5–15 min. Cells were then put in ice and the fluorescence measured with a BD FACSCalibur flow cytometer (BD) after Annexin V binding buffer was added to the mixture. The analysis tool for determining cell apoptosis is available from Cell Quest software (Becton Dickinson, Franklin Lakes, NJ).
Western Blot Analysis
Total proteins were extracted from breast tumor cells with RIPA lysis buffer. Separate equivalent amounts of proteins were obtained by running 12% SDS-PAGE gels and transferring targeted protein from the gel onto the PVDF membranes. When the above procedure was finished, the membrane was put into the mixture solution immersed in primary antibodies (1:1000 dilution) and incubated overnight at 4°C. The membrane was then incubated in secondary HRP rabbit IgG antibodies (1:2000 dilution) at room temperature for 1 h. The membrane was washed three times with TBST and targeted protein visualized using SuperSignal West Dura Extended Duration Substrate following the manufacturer’s instructions.
Statistical Analysis
The statistical analysis was performed using SPSS 20.0 software (IBM, SPSS, Chicago, IL) and GraphPad Prism (GraphPad, La Jolla, CA). Comparisons between groups were analyzed using Student’s t-tests. Kaplan-Meier plots and Log rank tests were used for evaluation of OS curves. Unless otherwise indicated, the data are presented as the mean ± S.D. of three independent experiments. P < 0.01 was considered statistically significant.
Results
Difference of Circ_0003645 Expression Across Breast Cancer Tissues
We applied GEO database GSE101123 to identify the expression of circ_0003645 in the clinical samples from the breast cancer patients. The results indicated that the expression of circ_0003645 in the cancerous tissues was higher than that in the matched normal tissues (Figure 1A).
Figure 1.
Up-regulated circ_0003645 in breast cancer tissues and cell lines. (A) Identification of circ_0003645 expression by GEO database analysis. (B and C) The expression of circ_0003645 in breast cancer tissues and cell lines. (D) The expression of circ_0003645 in two breast cancer cell lines after RNase R treatment. (E and F) Detection of circ_0003645 localization in breast cancer cells. **P < 0.01, ***P < 0.001, ##P < 0.01.
Among the 45 pairs of breast cancer tissue candidates, qRT-PCR analysis showed high expression of circRNA_0003645 in tumor tissues compared with corresponding normal tissues (P < 0.001; Figure 1B). In addition, we investigated the effect of the presence of circRNA_0003645 in 5 breast cancer cell lines (T47D, MCF-7, BT549, Hs-578T and MDA-MB-231) and normal breast epithelial MCF-10A and MCF-12A cells by validation with qRT-PCR using outward primer pairs. The results demonstrated higher expression of circRNA_0003645 in all the cancer cell lines than in normal breast epithelial MCF-10A cells, suggesting that the elevated level of circRNA_0003645 might be general to tumorigenesis of this cancer (P < 0.001; Figure 1C).
As shown in Figure 1C, the two cell lines with the highest expression of circRNA_0003645 in breast cancer cell lines were MCF-7 and MDA-MB-231, so we performed omitting reverse transcriptase reaction to verify circRNA_0003645 in these two selected cell lines by treating with RNase R prior to reverse transcription to confirm the structural lack of free 5- and 3- ends. The levels of linear transcript with GAPDH mock group demonstrated the efficacy of RNase R treatment. qRT-PCR results showed decreasing expression of GAPDH after RNase R treatment, while there was no significant change of circ_0003645 expression (Figure 1D).
Cellular localization analysis in cell lines (MCF-7 and MDA-MB-231) which exhibited much higher circRNA_0003645 expression revealed that circRNAs mainly resided in the cytoplasm (U6 as positive control for nuclear fraction; GAPDH for cytoplasmic fraction), a phenomenon that was consistent with the property of exonic circRNAs (Figure 1E and F). The assessment of the content of circRNA_0003645 in both two cell lines demonstrated the consistent results.
Circ_0003645 Promotes Tumor Cell Proliferation
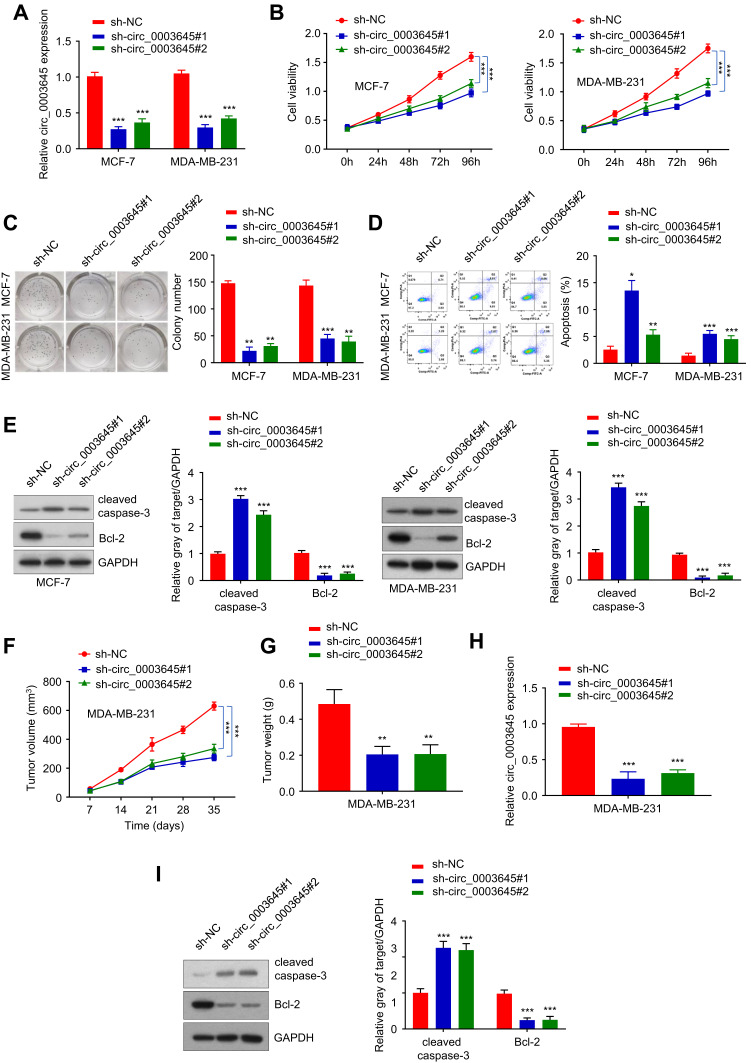
To investigate the role of circRNA_0003645 in breast cancer progression, we designed two siRNA oligonucleotides to target the unique back-splice junction in two breast cancer cell lines (MCF-7 and MDA-MB-231) to construct two stable knockdown cell strains (sh-circ_3645#1 and sh-circ_3645#2). qRT-PCR analysis provided knockdown efficiency: compared with the sh-NC control group, both two knockdown groups (sh-circ_0003645#1 and sh-circ_0003645#2) exhibited lower circ_0003645 expression (P < 0.01) and no effect in sh-NC group, indicating that the back-splice junction-specific siRNA successfully knocked down circRNA_0003645 (Figure 2A).
Figure 2.
Knockdown circ_0003645 in two breast cancer cell lines. (A) The expression of knockdown of circ_0003645 in MCF-7 and MDA-MB-231 cell lines using qRT-PCR detection efficiency. (B) CCK-8 analysis to assess cell proliferation after circ_0003645 inhibition. (C) Operation of colony formation assay to monitor cell colony-forming ability (left), and quantification of the colony numbers by Image (right). (D) Flow cytometry detecting cell apoptosis levels for suppressing circ_0003645 in MCF-7 and MDA-MB-231 cell lines. (E) Western Blot analysis showed protein levels changing related with cell apoptosis for suppressing circ_0003645 in MCF-7 and MDA-MB-231 cell lines. (F and G) Calculation of tumor weights after circ_0003645 knockdown. (H) qRT-PCR results exhibited knockdown circ_0003645 expression in tumors in two cell lines. (I) Western blot analysis showed cell apoptosis protein levels of sh-circ_0003645 in distinct subcutaneous tumor groups. *P < 0.05, **P < 0.01, ***P < 0.001.
For assessment of cell viability, we applied cell counting analysis (CCK-8) to detect the 450nm light absorption value in the three groups (sh-NC and sh-circ_0003645#1, sh-circ_0003645#2) in two cell lines (MCF-7 and MDA-MB-231 cells) at 0 h, 24 h, 48 h, 72 h and 96 h. Due to the specific knockdown, the cell numbers reduced significantly in both two cell lines groups (sh-circ_0003645#1 and sh-circ_0003645#2_) compared with the NC group (Figure 2B), suggesting the suppression of cell proliferation mediation. The cloning formation experiment detected the changes in the cloning ability of MCF-7 and MDA-MB-231 cells in each group. Compared with NC group, cloning numbers showed evident decreasing trends in the two groups (Figure 2C). To further characterize the role of circRNA_0003645 in tumor proliferation, we measured the cell apoptosis in three groups; knockdown of this circular RNA increased the apoptosis level in both cell lines, indicating that circRNA_0003645 was a regulation factor of the breast cancer (Figure 2D). Consistent with this, we found silenced circRNA_0003645 enhanced a key cell apoptosis factor — the cleaved caspase-3 protein level dramatically, while it decreased another key tumor marker — Bcl-2 protein level in both cell lines (Figure 2E).
To investigate the potential clinical relevance of circRNA_0003645 in vivo, we first injected MDA-MB-231 cells with or without circRNA_0003645 knockdown subcutaneously into dorsal flanks of model mice to construct subcutaneous tumor models and allowed them to proliferate for 35 days. With construction of breast tumor models from 3 stable cell strains (sh-NC, sh_circ_0003645#1 and sh-circ_0003645#2), we calculated tumor volumes with 7-day intervals. It was shown that the knockdown groups (sh_circ_0003645#1 and sh-circ_0003645#2) exhibited evidently smaller tumor volumes and lower tumor weights than those of the NC group at each 7 days in MDA-MB-231 cell line (P < 0.01, Figure 2F and G). Detection of sh_circ_0003645 expression in tumors showed decreased protein levels in knockdown groups (P < 0.01, Figure 2H). Compared with NC group, cleaved caspase-3 protein level was increased in subcutaneous tumor tissues and Bcl-2 expression was decreased in the knockdown groups (P < 0.01; Figure 2I).
Circ_0003645 May Serve as a miRNA Sponge
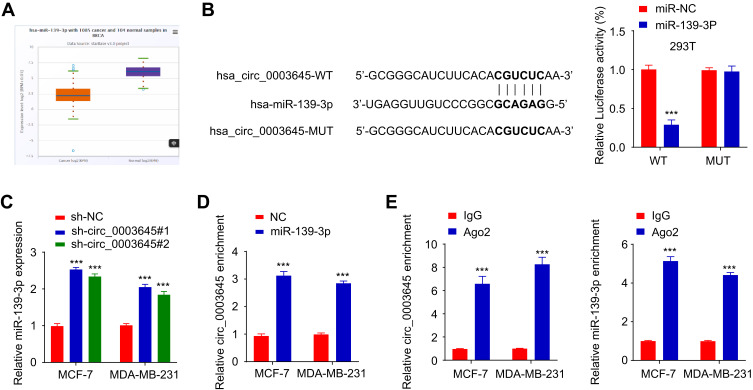
To explore the molecular function of circ_0003645, we performed a miRNA target sequence analysis using the Cancer Genome Atlas (TCGA) database. Sequencing data revealed that miR-139-3p was under-expressed in breast cancer tissues (Figure 3A). CircInteractome database analysis demonstrated that miR-139-3p targeting circ_0003645 contained several binding sites (Figure 3B). This caused us to hypothesize that circ_0003645 may serve as a miRNA sponge to protect a specific group of oncogenes from attacks by miRNAs in breast cancer. Next, we performed a luciferase reporter gene system conducted in 293T cells to test if the circ_0003645-miR-139-3p regulatory network is a critical component for oncogene activation. Over-expression of miR-139-3p could inhibit luciferase activity in cells, and the inhibitory effect disappeared after mutation of the predicted binding site of circ_0003645 (Figure 3B). Coherently, after lowering circ_0003645 in two cell lines (MCF-7 and MDA-MB-231), qRT-PCR results suggested that knockdown groups (sh_circ_0003645#1 and sh_circ_0003645#2) could enhance the miR-139-3p expression (Figure 3C).
Figure 3.
Circ_0003645 acts as a sponge for miR-139-3p. (A) The predicted expression of miR-139-3p in breast cancer proliferation through TCGA database. (B) The predicted binding sites of miR-139-3p within the circ_3645 sequence and luciferase assay of cells co-transfected with miR-139-3p mimics and wild-type or mutant luciferase reporter. (C) The expression of miR-139-3p after knockdown of circ_0003645 in MCF-7 and MDA-MB-231 cell lines using qRT-PCR analysis. (D) RNA pull-down experiment with miR-139-3p probes in two breast cancer lines (MCF-7 and MDA-MB-231). (E) RIP assay exhibiting the enrichment of circ_0003645 and miR-139-3p in the Ago2 fraction related with IgG fraction. ***P < 0.001.
To further consolidate the role of circ_0003645 as a miR-139-3p sponge, we developed an RNA pull down assay — a novel in vitro transcription combined with in vivo circularization method to test circ_0003645’s miRNA sponge effect in cells. Results demonstrated that the miR-139-3p probe enriched more circ_0003645 than the NC probe (Figure 3D). In addition, we applied RNA immuno-precipitation (RIP)-qRT-PCR assay and found more circ_0003645 and miR-139-3p enriched in the Ago2 group than in the IgG group (Figure 3E).
Circ_0003645 Promoted Breast Cancer Progression by Sponging miR-139-3p
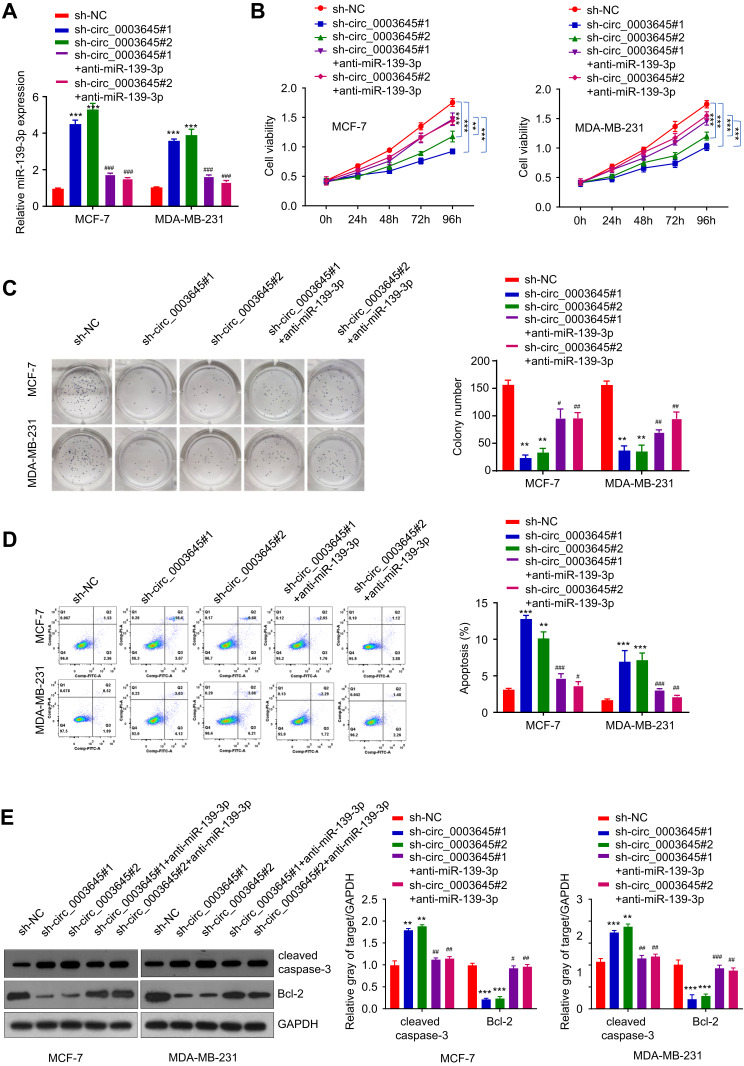
To further investigate the potential clinical relevance of cir_0003645 with sponging miR-139-3p in vivo, we detected the expression of miR-139-3p with knockdown of circ_0003645 in breast cancer cell lines (MCF-7 and MDA-MB-231). Compared with the NC group, the expression of miR-139-3p exhibited an increasing trend in both circular RNA knockdown groups (Figure 4A). Consistently, with miR-139-3p inhibitor transfection, the expression of miR-139-3p presented decreasing trend correspondingly (Figure 4A).
Figure 4.
Enhancement of breast cancer cell proliferation with circ_0003645 sponging miR-139-3p. (A) qRT-PCR experiments detecting miR-139-3p expression with or without inhibiting circ_0003645 or with or without suppressing miR-139-3p in two breast cancer cell lines. (B) CCK-8 analysis shown light absorption values at 450 nm in different time with or without inhibiting circ_0003645 and miR-139-3p in two breast cancer cell lines. (C) Colony formation assay detecting cloning numbers with or without inhibiting circ_0003645 or with or without suppressing miR-139-3p in two breast cancer cell lines. (D) Flow cytometry detecting cell apoptosis levels with or without inhibiting circ_0003645 or with or without suppressing miR-139-3p in two breast cancer cell lines. (E) Western blot analysis showing protein levels changing related with cell apoptosis with or without inhibiting circ_0003645 or with or without suppressing miR-139-3p in two breast cancer cell lines. **P < 0.01, ***P < 0.001, #P < 0.05, ##P < 0.01, ###P < 0.001.
To assess the cell viability under knockdown cir_0003645 and to further elucidate the cell growth situation with miR-139-3p inhibition, we performed CCK-8 analysis to detect OD values at 450nm at fixed interval periods (0 h, 24 h, 48 h, 72 h and 96 h) in MCF-7 and MDA-MB-231 cells. Our results showed that two knockdown circ_0003645 RNA groups exhibited reduced OD values compared with the NC group at each time (Figure 4B). Meanwhile, transfection of anti-miR-139-3p into these groups could reverse this situation (P < 0.01, Figure 4B). Cloning formation analysis in both circular RNA knock-down groups (sh-circ_0003645#1 and sh-circ_0003645#2) showed far fewer numbers than those of the control group (Figure 4C), suggesting cir_0003645 is an important regulatory factor in breast cancer cells. With anti-miR-139-3p transfecting into two knockdown circ_0003645 cell lines, cell cloning ability presented partial recovery with increasing cloning numbers (P < 0.01; Figure 4C). The specific knockdowns in two circ_0003645 groups led to a dramatic rise in the apoptosis levels in both cells lines (Figure 4D). While suppression of miRNA-139-3p could neutralize their ascending trends (P < 0.01; Figure 4D). To evaluate apoptosis-related protein levels of two circ_0003654 knockdown groups (sh_circ_0003645#1, sh-circ_0003645#2), we tested apoptosis-related caspase-3 expression and the breast cancer marker Bcl-2 expression. Results demonstrated two knockdown groups (sh_circ_0003645 #1 or sh-circ_0003645#2) showed increased cleaved caspase-3 protein levels and decreased Bcl-2 protein levels, indicating inhibiting circ_0003645 may serve as a regulatory function in breast cancer progression. But, when we transfected depressed miR-139-3p into the above cells, results showed that it could partially decrease cleaved caspase-3 protein levels and partially elevate the Bcl-2 protein levels (P < 0.01, Figure 4E)
Circ_0003645 Positively Regulated HMGB1 by Sponging miR-139-3p
Using Targescan software, we predicted the presence of the miR-139-3p binding sites in the 3ʹ non-coding region of HMGB1. Over-expression of miR-139-3p could inhibit the luciferase activity in 293T cells. After mutation in the predicted HMGB1 3ʹ-UTR binding site, its inhibitory function disappeared (Figure 5A and B).
Figure 5.
Circ_0003645 regulates HMGB1 by sponging miR-139-3p. (A and B) Prediction of binding sites of miR-139-3p within HMGB sequence and luciferase assay of cells co-transfected with miR-139-3p mimics and wild-type or mutant luciferase reporter in 293T cells. (C and D) mRNA and protein expressions of HMGB1 under over-expression miR-139-3p in MCF-7 and MDA-MB-231 cell lines. (E and F) mRNA and protein expressions of HMGB1 in with or without inhibiting circ_0003645 groups or with or without suppressing miR-139-3p groups. **P < 0.01, ***P < 0.001, #P < 0.05, ##P < 0.01, ###P < 0.001.
We also detected the mRNA and protein expression levels of HMGB1 in MCF-7 and MDA-MB-231 cells with miR-139-3p over-expression. Results demonstrated that miR-139-3p over-expression reduced the mRNA and protein expressions of HMGB1. qRT-PCR and Western blot analysis also provided HMGB1 mRNA and protein expressions in the different groups (sh-NC, sh_circ_0003645#1, sh-circ_0003645#2, sh_circ_0003645#1) in MCF-7 and MDA-MB-231 cells (Figure 5C and D). Comparison between vehicle control and circular RNA knockdown groups generated sh_circ_0003645 #1 or sh-circ_0003645#2 reduced HMGB1 mRNA and protein expression. With inhibited circular RNA knockdown by adding anti-miR-139-3p, two groups exhibited partial increase of the expression levels of HMGB1 mRNA and protein (P < 0.01, Figure 5E and F).
Discussion
CircRNAs represent a class of RNAs with notable particularities regarding their architecture, abundance, and expression patterns in different cell types and pathologies.30 The expression of circRNAs in human diseases is an intensely researched topic. Previous research showed that the molecular characters of circRNA among distinct cancer cell lines were highly diverse compared with normal cell lines.31 Despite a few earlier studies reporting that circRNAs are down-regulated in cancer cells,32–34 there are increasing recent studies showing an enrichment of certain circRNAs in several types of human tumors.35–39 Although a well-characterized circ_0003645 study displayed the same behavior with elevated expression in non-small cell lung cancer tissue and cell lines,21 there was no other study available for circ_0003645 in the breast cancer field. In our study, using GEO database prediction, we found the expression of a circular RNA (circ_0003645) enriched in breast tumor specimens. Our data demonstrated that this circular RNA upregulated in all stages of breast cancer and negatively correlated with prognosis. In vitro and in vivo studies showed that circ_0003645 possessed oncogenic capability in terms of promoting cell proliferation.
Previous research revealed that circRNA acts as a sponge of microRNA, which results in the loss of miRNA function, accompanied by an increased gene target.40,41 As an essential class of non-coding small RNAs, microRNAs (miRNAs) could control target gene expression at the post-transcriptional level and play a critical role in the pathological processes,42 which can be as a non-invasive biomarker for colorectal cancer,43 an onco-suppressor for bladder cancer, cervical cancer and ovarian cancer.25,44,45 Unusual expression of miRNAs is closely correlative to the development of glioma, which provides a potential therapeutic target for this disease.46 Several studies reported that miR-139-3p was under-expressed in many cancers, such as colorectal cancer, head and neck cancer, and hepatocellular carcinoma, which possibly served as a tumor inhibitor.47,48 Other research showed that miR-139-3p could target MMP11 to suppress bladder cancer invasion and metastasis.44 Some experts confirmed the key function of miR-139-3p in HPV remedy by exhibiting its inhibition of HPV-16 proteins and recovering key tumor suppressor proteins p53, p16, and p21, thereby preventing HPV-16 positive cells progression and migration.49 Nevertheless, which miR-139-3p regulates the development of breast cancer is largely unknown. In this study, through bioinformatics analysis, we found that circ_0003645 contained potential binding sites to the miR-139-3p sequence. With miR-139-3p over-expression, circ_0003645 expression was downregulated in breast cell lines and tissues.
HMGB1 serves a critical role in inflammatory diseases. With specific capability for release into the extracellular environment, HMGB1 is not only a non-histone nuclear protein that can adjust gene transcription,50 but also can be passively migrated into the extracellular space as a pro-inflammatory mediator by necrotic cells, or in an active manner by lipopolysaccharide (LPS)-, tumor necrosis factor-α (TNF-α)-, interleukin-1β- and interferon-γ-stimulated monocytes or macrophages.51 Previous studies demonstrated that the extracellular functions of HMGB1 are dependent on its redox state and combination of target receptors, which regulate the responses to inflammation, immunity, chemotaxis and tissue regeneration.52–54
Extracellular HMGB1 is not only a critical mediator of lethality in inflammatory diseases,55 but it also plays a vital role in breast cancer. Several studies have shown that upregulation of HMGB1 can lead to proliferation, migration and invasion of breast cancer cells. For instance, miR-200c inhibits metastasis of breast cancer cells by targeting HMGB1.56 HMGB1 promotes cancer cell migration and invasion, as well as angiogenesis, growth and metastasis of cancer by regulating matrix metallopeptidase 9 (MMP‑9) in lung cancer.57 In the present study, we reported that by acting as a “miR-139-3p sponge“, circ_0003645 could positively mediate HMGB1 expression in breast cancer cells and tissues.
In conclusion, our study demonstrates that circ_0003645 can enhance proliferation or induce apoptosis of breast cancer cells by regulation of miR-139-3p/HMGB1. Circ_0003645 might act as a “miR-139-3p sponge” and appears to be a new therapeutic target for the treatment of breast cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Anastasiadi Z, Lianos GD, Ignatiadou E, et al. Breast cancer in young women: an overview. Updates Surg. 2017;69(3):313–317. doi: 10.1007/s13304-017-0424-1 [DOI] [PubMed] [Google Scholar]
- 2.Fahad Ullah M. Breast cancer: current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51–64. [DOI] [PubMed] [Google Scholar]
- 3.Moon PG, Lee JE, Cho YE, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. 2016;22(7):1757–1766. doi: 10.1158/1078-0432.CCR-15-0654 [DOI] [PubMed] [Google Scholar]
- 4.Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012;6(2):140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuk K, Zucknick M, Heil J, et al. Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer. 2013;132(7):1602–1612. doi: 10.1002/ijc.27799 [DOI] [PubMed] [Google Scholar]
- 6.Inns J, James V. Circulating microRNAs for the prediction of metastasis in breast cancer patients diagnosed with early stage disease. Breast. 2015;24(4):364–369. [DOI] [PubMed] [Google Scholar]
- 7.Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du W, Li D, Guo X, et al. Circ-PRMT5 promotes gastric cancer progression by sponging miR-145 and miR-1304 to upregulate MYC. Artif Cells Nanomed Biotechnol. 2019;47(1):4120–4130. doi: 10.1080/21691401.2019.1671857 [DOI] [PubMed] [Google Scholar]
- 12.Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 14.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 15.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Huang C, Bao C, et al. Corrigendum: exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2017;24(2):194. doi: 10.1038/nsmb0217-194a [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Dong Y, Zhao L, et al. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53(4):1752–1762. [DOI] [PubMed] [Google Scholar]
- 19.Huang G, Li S, Yang N, et al. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 20.Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027 [DOI] [PubMed] [Google Scholar]
- 21.An J, Shi H, Zhang N, et al. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;511(4):921–925. doi: 10.1016/j.bbrc.2019.03.011 [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Yuan Y, Li HY. MicroRNA-139-3p suppresses growth and metastasis of glioblastoma via inhibition of NIN1/RPNI2 binding protein 1 homolog. Eur Rev Med Pharmacol Sci. 2019;23(10):4264–4274. [DOI] [PubMed] [Google Scholar]
- 23.Tian W, Wu W, Li X, et al. MiRNA-139-3p inhibits the proliferation, invasion, and migration of human glioma cells by targeting MDA-9/syntenin. Biochem Biophys Res Commun. 2019;508(1):295–301. doi: 10.1016/j.bbrc.2018.11.144 [DOI] [PubMed] [Google Scholar]
- 24.Zou ZC, Dai M, Huang ZY, et al. MicroRNA-139-3p suppresses tumor growth and metastasis in hepatocellular carcinoma by repressing ANXA2R. Oncol Res. 2018. doi: 10.3727/096504018X15178798885361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang P, Xi J, Liu S. MiR-139-3p induces cell apoptosis and inhibits metastasis of cervical cancer by targeting NOB1. Biomed Pharmacother. 2016;83:850–856. doi: 10.1016/j.biopha.2016.07.050 [DOI] [PubMed] [Google Scholar]
- 26.Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. doi: 10.1038/s41419-019-1382-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng H, Gu ZY, Li Q, et al. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J Ovarian Res. 2019;12(1):35. doi: 10.1186/s13048-019-0508-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24(1):40. doi: 10.1186/s10020-018-0039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi: 10.1016/j.cell.2018.03.059 [DOI] [PubMed] [Google Scholar]
- 30.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025. [PMC free article] [PubMed] [Google Scholar]
- 34.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. doi: 10.3233/CBM-150552 [DOI] [PubMed] [Google Scholar]
- 35.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 36.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;166(4):1055–1056. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed I, Karedath T, Andrews SS, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7(24):36366–36381. doi: 10.18632/oncotarget.8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7(30):47186–47200. doi: 10.18632/oncotarget.9706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sand M, Bechara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83(3):210–218. doi: 10.1016/j.jdermsci.2016.05.012 [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Hou J, He D, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32(4):211–224. doi: 10.1016/j.tig.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol. 2019;234(8):13807–13819. [DOI] [PubMed] [Google Scholar]
- 42.Huo LW, Wang YF, Bai XB, et al. circKIF4A promotes tumorogenesis of glioma by targeting miR-139-3p to activate Wnt5a signaling. Mol Med. 2020;26(1):29. doi: 10.1186/s10020-020-00159-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ng L, Wan TM, Man JH, et al. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8(16):27393–27400. doi: 10.18632/oncotarget.16171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yonemori M, Seki N, Yoshino H, et al. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107(9):1233–1242. doi: 10.1111/cas.13002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue F, Li QR, Xu YH, et al. MicroRNA-139-3p inhibits the growth and metastasis of ovarian cancer by inhibiting ELAVL1. Onco Targets Ther. 2019;12:8935–8945. doi: 10.2147/OTT.S210739 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Zhou Q, Liu J, Quan J, et al. MicroRNAs as potential biomarkers for the diagnosis of glioma: a systematic review and meta-analysis. Cancer Sci. 2018;109(9):2651–2659. doi: 10.1111/cas.13714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanaan Z, Roberts H, Eichenberger MR, et al. A plasma microRNA panel for detection of colorectal adenomas: a step toward more precise screening for colorectal cancer. Ann Surg. 2013;258(3):400–408. doi: 10.1097/SLA.0b013e3182a15bcc [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Gao J, Zhou B, et al. Identification of prognostic markers for hepatocellular carcinoma based on miRNA expression profiles. Life Sci. 2019;232:116596. doi: 10.1016/j.lfs.2019.116596 [DOI] [PubMed] [Google Scholar]
- 49.Sannigrahi MK, Sharma R, Singh V, et al. Role of host miRNA Hsa-miR-139-3p in HPV-16-induced carcinomas. Clin Cancer Res. 2017;23(14):3884–3895. doi: 10.1158/1078-0432.CCR-16-2936 [DOI] [PubMed] [Google Scholar]
- 50.Ueda T, Chou H, Kawase T, et al. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry. 2004;43(30):9901–9908. doi: 10.1021/bi035975l [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Bloom O, Zhang M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285(5425):248–251. doi: 10.1126/science.285.5425.248 [DOI] [PubMed] [Google Scholar]
- 52.Janko C, Filipovic M, Munoz LE, et al. Redox modulation of HMGB1-related signaling. Antioxid Redox Signal. 2014;20(7):1075–1085. doi: 10.1089/ars.2013.5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang D, Kang R, Zeh HJ, et al. High-mobility group box 1, oxidative stress, and disease. Antioxid Redox Signal. 2011;14(7):1315–1335. doi: 10.1089/ars.2010.3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venereau E, Casalgrandi M, Schiraldi M, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 2012;209(9):1519–1528. doi: 10.1084/jem.20120189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersson U, Tracey KJ. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol. 2011;29:139–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang BP, Wang DS, Xing JW, et al. miR-200c inhibits metastasis of breast cancer cells by targeting HMGB1. J Huazhong Univ Sci Technol Med Sci. 2014;34(2):201–206. doi: 10.1007/s11596-014-1259-3 [DOI] [PubMed] [Google Scholar]
- 57.Wu L, Yang L. The function and mechanism of HMGB1 in lung cancer and its potential therapeutic implications. Oncol Lett. 2018;15(5):6799–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]