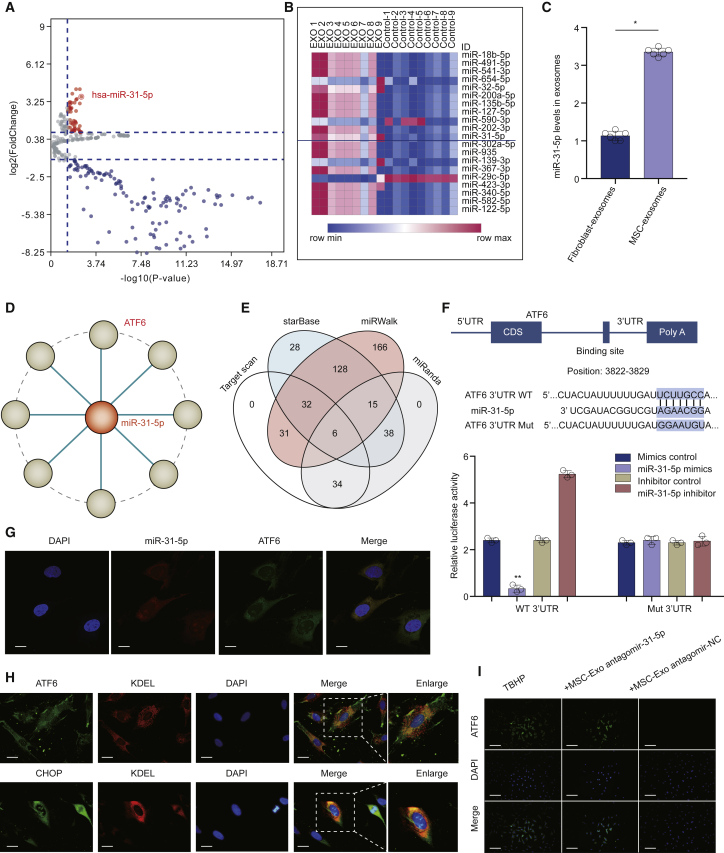
Figure 4.
miR-31-5p Was Highly Expressed in MSC-Exosomes, and ATF6 Was a Target of miR-31-5p
(A) Volcano plots depicted differential expression of miRNAs examined using miRNA microarray in MSC-exosomes compared to cells. (B) Heatmap of 20 upregulated miRNAs in microarray. (C) qRT-PCR assay verified the upregulation of miR-31-5p in MSC-exosomes compared to fibroblast-exosomes. (D) Cystoscope was utilized to verify the targets of miR-31-5p. (E) Venn diagram indicated the targets using different algorithms. (F) EPCs were inserted with miR-31-5p and luciferase constructs of ATF6 with the wild-type-putative miR-31-5p binding sites or mutated sites via transfection. (G) FISH revealed that both miR-31-5p and ATF6 mRNA were localized in the cytoplasm. Blue fluorescence designates the nucleus, red fluorescence designates miR-31-5p, and green fluorescence designates ATF6 mRNA (scale bars, 10 μm). (H) Immunofluorescence double staining for colocalization of ATF6 and CHOP with KDEL were localized in the endoplasmic reticulum (ER) of EPCs (scale bars, 20 μm). (I) Immunofluorescence of ATF6 protein in EPCs (scale bars, 100 μm). Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.