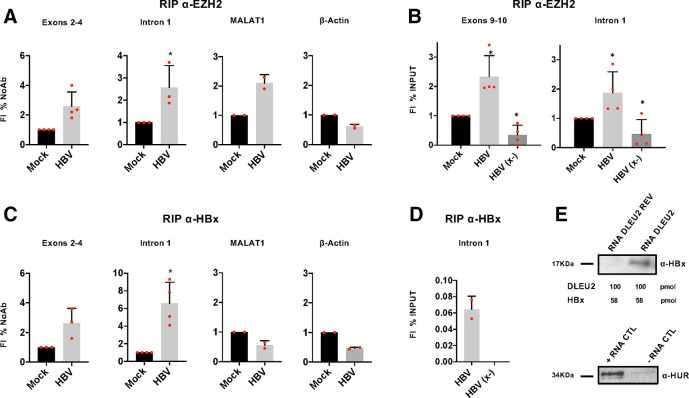
Figure 2.
DLEU2 long non-coding RNA (lncRNA) interacts with enhancer of zeste homolog 2 (EZH2) and directly binds HBx. (A) RNA binding protein immunoprecipitation (RIP) using anti-EZH2 antibody. Nuclear lysates of mock-infected and HBV-infected primary human hepatocytes (PHHs) (72 hours) were subjected to immunoprecipitation with anti-EZH2 antibody or control IgG. DLEU2 exons 2–4 or intron 1, MALAT1 (positive control) and β-actin (negative control) were detected by PCR and analysed by relative densitometry. (B) RIP using anti-EZH2 antibody. Nuclear lysates of mock, HBV-infected and HBV (x-)-infected PHHs (72 hours) were subjected to immunoprecipitation with anti-EZH2 antibody or control IgG. DLEU2 exons 9–10 or intron 1 were detected by quantitative PCR (qPCR) assay. RIP results are expressed as fold induction (FI) of the % of input with respect to mock. (C) RIP using anti-HBx antibody. Nuclear lysate of mock-infected and HBV-infected PHHs (72 hours) were subjected to immunoprecipitation with anti-HBx antibody or control IgG. DLEU2 exons 2–4 or intron 1, MALAT1 and β-actin (negative controls) were detected by PCR assay and analysed by densitometry relative to the mock. RIP data in (A) and (B) represent means±SD from at least three independent experiments. (D) RIP using anti-HBx antibody. Nuclear lysate of mock, HBV-infected and HBV (x-)-infected PHHs (72 hours) were subjected to immunoprecipitation with anti-HBx antibody or control IgG. Intron 1 was detected by real-time qPCR assay. RIP results are expressed as % of input after subtraction of mock. (E) Immunoblot analysis of RNA pull-downs showing the interaction between the DLEU2 RNA (100 pmol of 3’-desthiobiotinylated RNA oligonucleotide) and recombinant HBx (1 µg), using a DLEU2 reverse sequence (DLEU2 REV) RNA as negative control (upper panel). Immunoblot analysis of positive and negative controls (provided by the manufacturer), which respectively contain RNA regions with or without binding sites for HUR, a RNA-binding proteins that regulates mRNA stability (lower panel). Data from one representative experiment out of three independent experiments are shown. In panels (A) and (B), *p<0.05; Mann-Whitney U test.