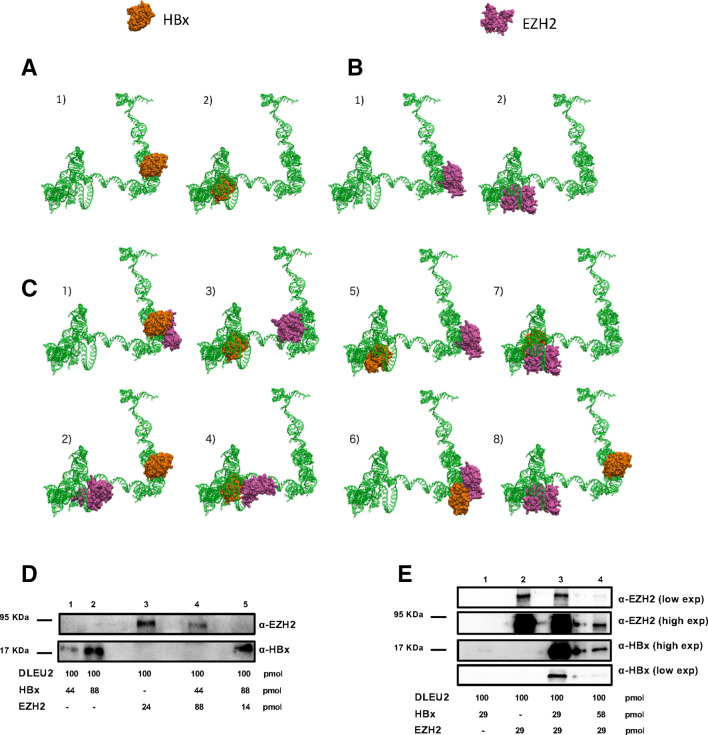
Figure 3.
HBx and enhancer of zeste homolog 2 (EZH2) directly bind DLEU2 and compete for partially overlapping sites. (A) Configurations obtained from docking of HBx (surface representation, orange) on three-dimensional (3D) structure of DLEU2 intron 1 (green). Panels 1–2 present the two most probable loci for HBx adsorption on DLEU2 intron 1. (B) Configurations obtained from docking of EZH2 (surface representation, mauve) on 3D structure of DLEU2 intron 1 (green). Panels 1–2 present the two most probable loci for EZH2 adsorption on DLEU2 intron 1. (C) Panels 1–2: two most probable configurations obtained from docking of EZH2 on the most probable HBx-DLEU2 intron 1 complex (panel (A)—1). Panels 3–4: two most probable configurations obtained from docking of EZH2 on the second most probable HBx-DLEU2 intron 1 complex (panel (A)—2). Panels 5–6: two most probable configurations obtained from docking of HBx on the most probable EZH2-DLEU2 intron 1 complex (panel (B)—1). Panels 7–8: two most probable configurations obtained from docking of HBx on the second most probable EZH2-DLEU2 intron 1 complex (panel (B)—2). (D) Immunoblot analysis of RNA pull-downs showing the interaction between DLEU2 RNA, HBx and EZH2. HBx (0.75 µg=44 pmol or 1.5 µg=88 pmol) or EZH2 (1.2 µg=14 pmol, 2 µg=24 pmol or 7.5 µg=88 pmol) recombinant proteins alone or in combination (EZH2:HBx ratios of 2:1 or 1:6) were added to a fixed amount of DLEU2 RNA (100 pmol). (E) Immunoblot analysis of RNA pull-downs showing the interaction between DLEU2 RNA (100 pmol), HBx and EZH2. Increasing concentrations of HBx (0.5 µg=29 pmol, 1 µg = 58 pmol) were added at a fixed concentration of EZH2 (2.5 µg=29 pmol) recombinant proteins corresponding to EZH2:HBx ratios of 1:1 and 1:2, respectively. Data from one representative experiment out of three independent experiments are shown.