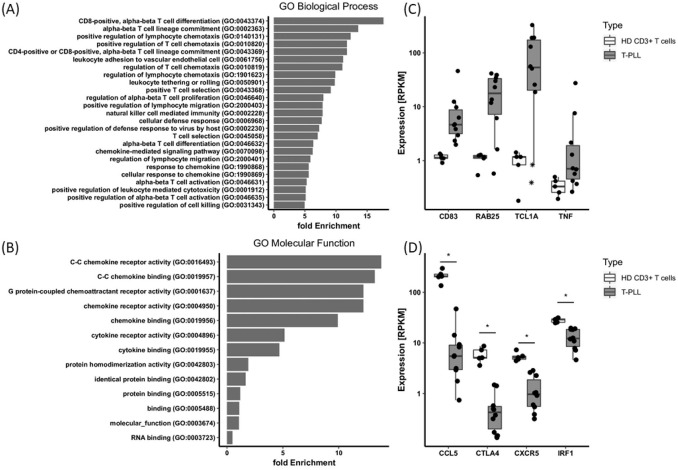
Figure 2.
Comparative transcriptomic profiling of T-cell prolymphocytic leukemia (T-PLL) and healthy donor (HD) T-cells. RNA sequencing expression data from ten T-PLL samples were compared with CD3+ T-cells from five HD yielding a panel of 674 differentially expressed genes (DEG, FC ⩾ 2 or FC ⩽ –2, p ⩽ 0.05, Supplemental Table S2). (A), (B), Fold enrichment of differential expressed genes in gene ontology (GO) categories biological process and molecular function as identified by GO enrichment analysis (false discovery rate < 0.5). For biological process, only the top 25 GO terms are displayed. (C), (D), mRNA expression levels of individual DEG, bars indicate the median and lower and upper hinges mark the 25th and 75th percentile. Whiskers range to the smallest or highest value, respectively, but not further than 1.5 × interquartile range. *p < 0.05 (Student’s t-test, corrected). Note, outlier in the TCL1A plot marked by an asterisk represent T-PLL cases 2 and 8 with an t(X;14) resulting in overexpression of MTCP1 (not shown) but lacking cytogenetic evidence of TCL1A rearrangement.
RPKM, reads per kilobase per million mapped reads.