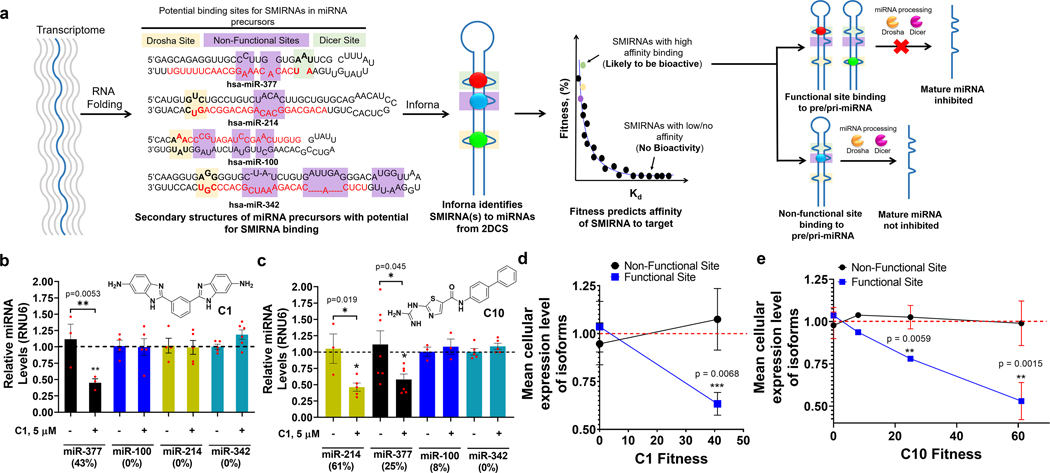
Fig. 2 |. SMIRNA strategy and method validation in vitro and in HUVECs.
a, Schematic of how SMIRNAs are identified from sequencing using Inforna. Features with high fitness scores are likely to yield bioactive interactions. For miRNAs, SMIRNAs must have sufficient fitness and occupy a functional site (Drosha site, tan boxes; Dicer site, green boxes; non-functional sites, purple boxes). pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA. b,c, Effect of C1 (b) and C10 (c) on miRNAs with different fitness scores in HUVECs, as determined by measuring mature miRNA levels by RT–qPCR. Data are reported as mean ± s.e.m. (n= 4 biologically independent samples). d,e, Activity of C1 (d) and C10 (e) as a function of fitness score against all isoforms of miR-377, miR-214, miR-100 and miR-342. Data are reported as mean ± s.e.m. In d, n= 15 miRNAs where the SMIRNA binds to non-functional sites and n= 2 miRs where the SMIRNA binds to functional sites; in e, n= 15 miRNAs where the SMIRNA binds to non-functional sites and n= 4 miRNAs where the SMIRNA binds to functional sites. All P values were determined from a two-sided Student’s t-test; *, P<0.05; **, P< 0.01; ***, P< 0.001.